Product details

NanOnCT Panel
#1001901 (96 rxn)
NanOnCT Panel v1.0 is a compactable panel designed for analysis of solid tumors and liquid biopsy. This panel involves a total number of 69 genes with selected regions covered. With an optimized probe design, this panel offers a highly uniform coverage over targeted regions for both FFPE and cell-free DNA.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Gene List
Genes with coding regions covered
Genes with full coding regions shown in bold.
AKT1
ALK
APC
AR
ARAF
ATM
BRAF
BRCA1
BRCA2
CCND1
CDH1
CDK12
CDK4
CDK6
CDKN2A
CTNNB1
DDR2
EGFR
ERBB2
ERCC2
ESR1
EZH2
FGFR1
FGFR2
FGFR3
GNA11
GNAQ
GNAS
HRAS
IDH1
IDH2
JAK2
JAK3
KDM6A
KIT
KRAS
MAP2K1
MAP2K2
MDM2
MET
MLH1
MPL
MSH2
MSH6
MTOR
MYC
NF1
NPM1
NRAS
NTRK1
NTRK2
NTRK3
PDGFRA
PIK3CA
PMS2
PTCH1
PTEN
PTPN11
RAF1
RB1
RET
ROS1
SMARCB1
SMO
STK11
TERT
TP53
TSC1
TSC2
Genes with selected introns covered
ALK
BRAF
EGFR
FGFR2
FGFR3
MET
NTRK1
NTRK2
PDGFRA
RET
ROS1
Microsatellite Markers
BAT-25
BAT-26
BAT-40
BAT-RII
NR-21
NR-22
NR-24
NR-27
MONO-27
D2S123
D5S346
D17S261
D17S520
D18S34
Performance
Table 1. On-target rate for gDNA and cfDNA samples
Platforms
gDNA
On-target rate (%)
cfDNA
On-target rate (%)
HiSeq X Ten, PE150
87.02
87.35
DNBSEQ-G400, PE100
85.47
87.00
The DNA libraries were prepared using NadPrep library kits with 50 ng of human genomic DNA (Promega, G1512) and 10 ng of plasma cfDNA from a healthy donor, respectively. 1 M read pairs were subsampled for the analysis. On-target rates were calculated as the percentage of mapped read pairs that overlap with probe regions.
Coverage Uniformity
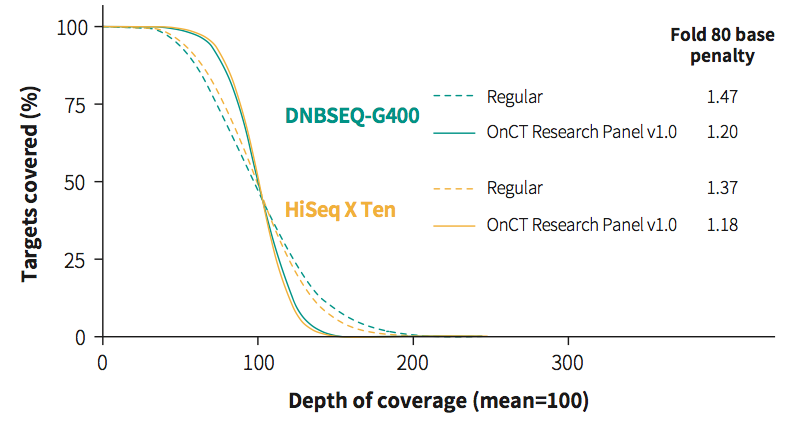
Fig 1. Improved design cfDNA capture. The optimized design of NanOnCT Panel v1.0 showed improved coverage uniformity for cfDNA libraries on both Illumina®️ and MGI platforms.
Variant Analysis
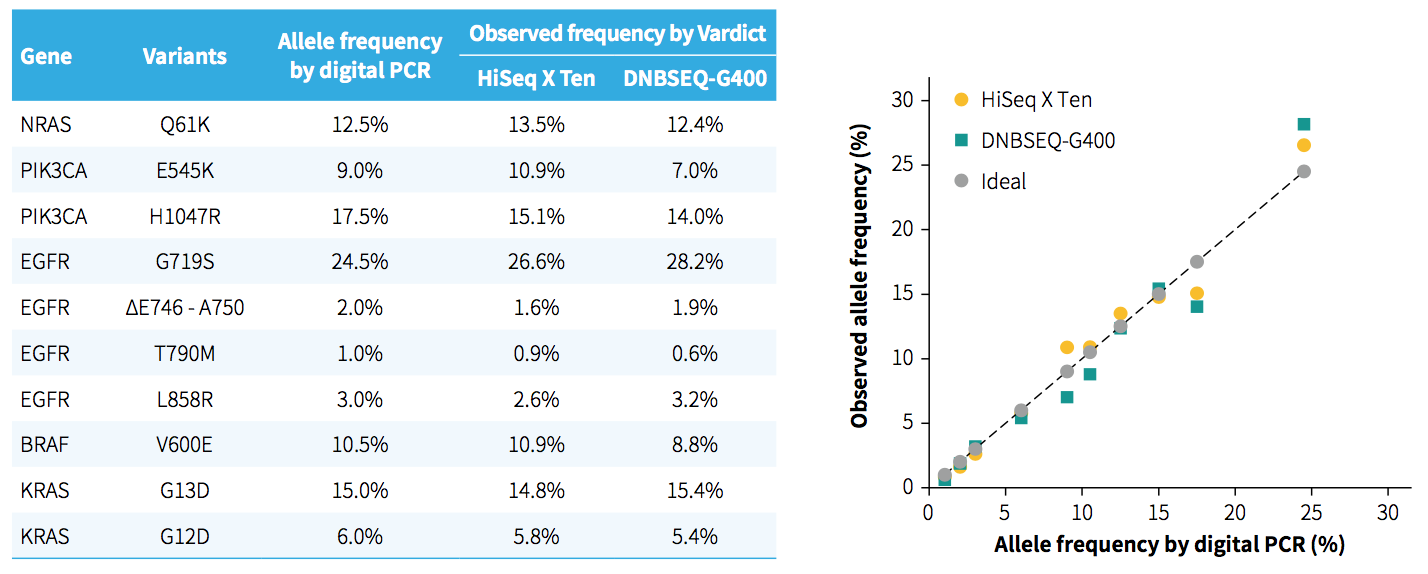
Fig 2. Evaluation of NanOnCT Panel v1.0 on the analysis of SNVs and indels. DNA libraries were prepared from Onco SNV Multiplex 1-25% gDNA (Genewell, GW-OGTM004) using NadPrep library kits. The enriched libraries were sequenced either on HiSeq X Ten (PE150) or DNBSEQ-G400 (PE100).
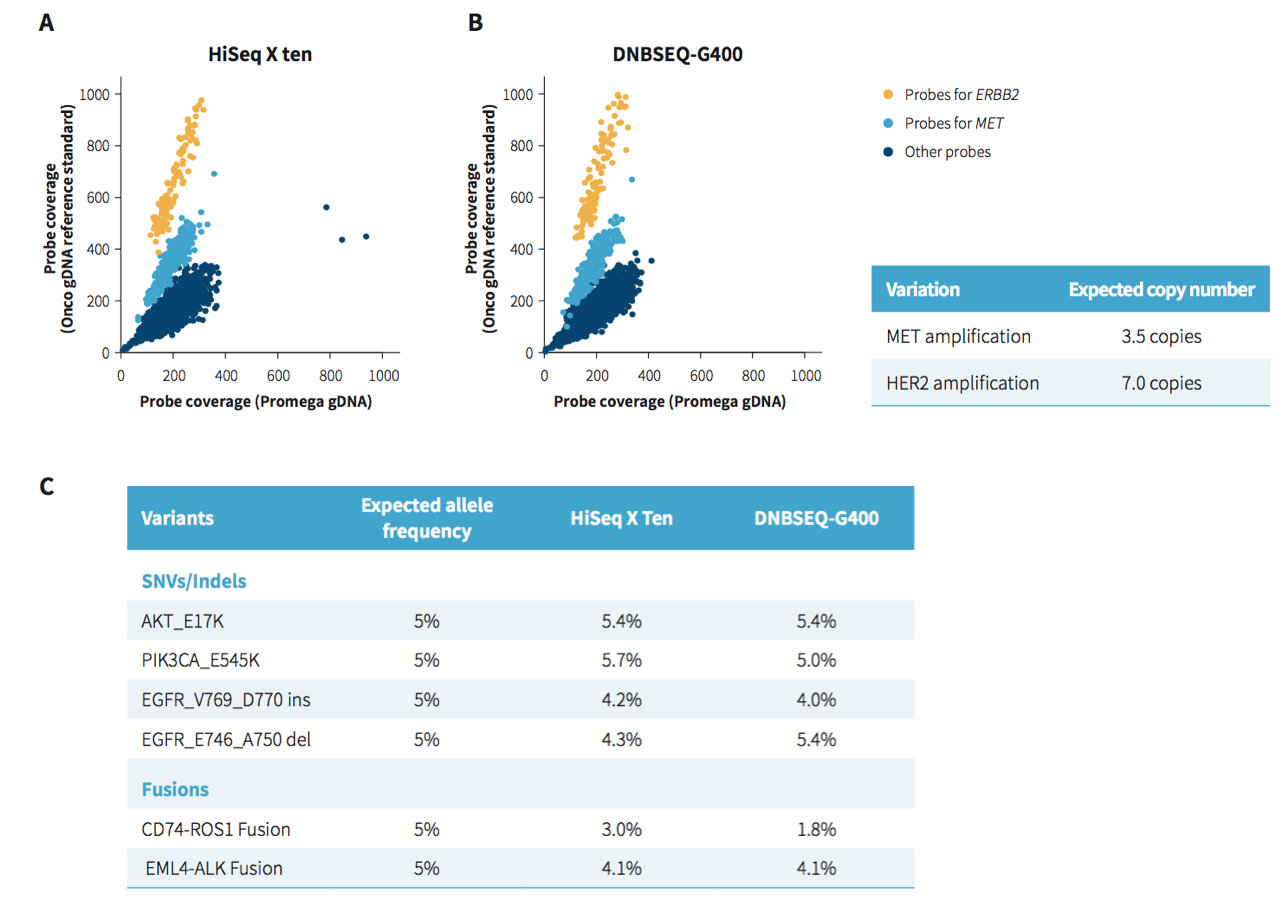
Fig 3. Evaluation of NanOnCT Panel v1.0 on the analysis of structural variations. A. & B. Gene amplification; C. SNVs with regions of high or low GC content and long insertion / deletion. The DNA libraries were prepared from Onco Structural Multiplex 5% gDNA (GeneWell, GW- OGTM001) using NadPrep library kits. The libraries were enriched using the NanOnCT Panel v1.0. The enriched libraries were sequenced either on HiSeq X Ten (PE150) or DNBSEQ-G400 (PE100).
Coverage Over Selected Microsatellite Markers

Fig 4. Effective coverage over microsatellite markers. The enriched libraries were sequenced on a HiSeq X Ten platform (PE150). Only reads spanning full-length of microsatellite markers were counted as effective reads.
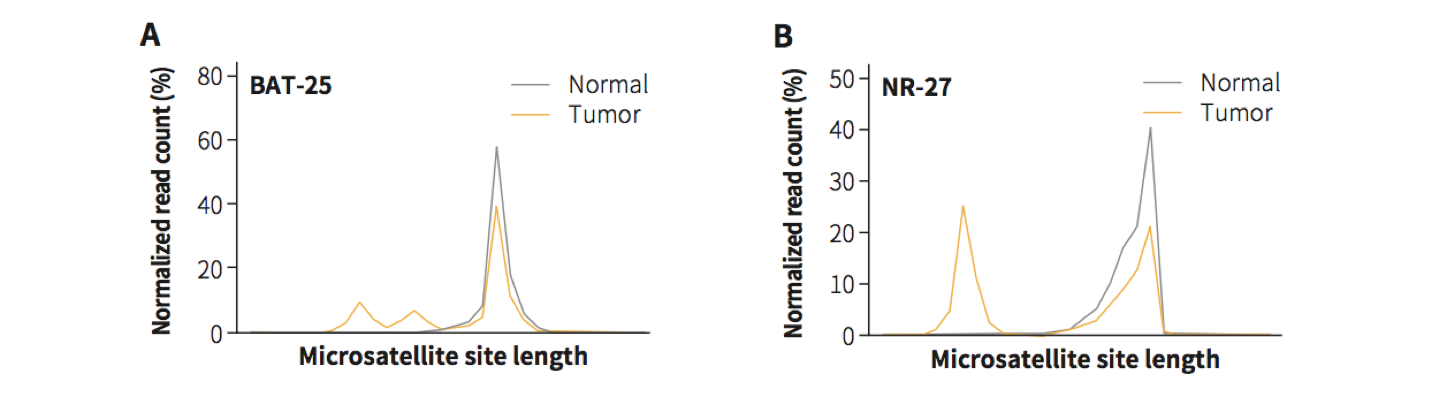
Fig 5. Length distribution of microsatellite site in a sample with microsatellite instability (MSI) for A. BAT-25 and B. NR-27.
For research use only. Not for use in diagnostic procedures.
|
Catalog# |
Color Of Tube Cap |
Item |
Volume |
Package/Storage |
|
1001902 |
|
NanOnCT Panel v1.0, 16 rxn |
70 μl×1 |
–20℃ |
|
1001901 |
|
NanOnCT Panel v1.0, 96 rxn |
415 μl×1 |
–20℃ |
|
Product |
Catalog# |
|
NanOnCT Panel v1.0, 16 rxn |
1001902 |
|
NanOnCT Panel v1.0, 96 rxn |
1001901 |
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com


