Product details

Ext-RNA Control Panel
#1001651 (96 rxn)
The ERCC RNA Spike-in Control Mixes is developed by National Institute of Standards and Technology (NIST) which is composed of a serious of RNA sequences with determined concentrations. It can derive standard curves relating read counts to RNA concentration and is widely used in performance assessment of gene expression experiments.
Ext-RNA Control Panel v1.0 targeted 24 of 92 ERCC transcripts 273-2,022 bp in length, with concentration ranging from 0.014305 attomol/μL to 937.5 attomol/μL. The 24 ERCC RNAs will serve as external controls for RNA expression quantification.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
List of Targeted 24 ERCC RNAs
Table 1. Information of targeted 24 ERCC RNAs.
ERCC ID
Concentration
(attomol/μL)
Length
(bp)
ERCC ID
Concentration
(attomol/μL)
Length
(bp)
ERCC ID
Concentration
(attomol/μL)
Length
(bp)
ERCC-00057
0.014305
1,021
ERCC-00058
1.831055
1,136
ERCC-00170
14.64844
525
ERCC-00017
0.114441
1,136
ERCC-00077
3.662109
743
ERCC-00084
29.29688
994
ERCC-00016
0.228882
844
ERCC-00150
3.662109
273
ERCC-00025
58.59375
1,994
ERCC-00156
0.457764
494
ERCC-00034
7.324219
844
ERCC-00071
58.59375
642
ERCC-00158
0.457764
1,027
ERCC-00085
7.324219
1,019
ERCC-00112
117.1875
1,136
ERCC-00109
0.915527
536
ERCC-00157
7.324219
1,019
ERCC-00092
234.375
1,124
ERCC-00137
0.915527
537
ERCC-00059
14.64844
1,023
ERCC-00042
468.75
1,023
ERCC-00033
1.831055
2,022
ERCC-00126
14.64844
1,118
ERCC-00108
937.5
1,022
Workflow

Fig 1. The workflow of RNA targeted capture sequencing.
Performance
ERCC Standard Validation
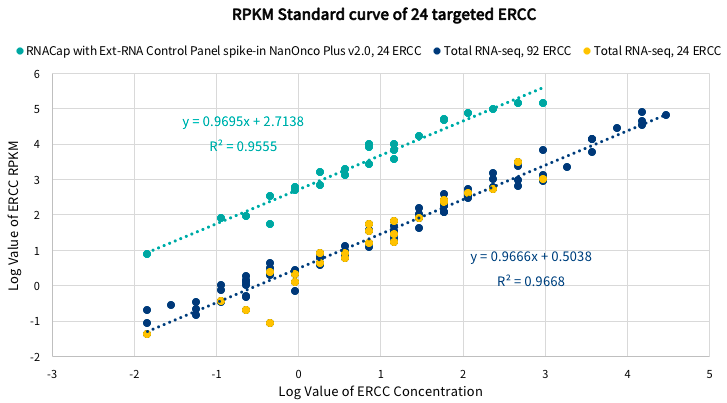
Fig 2. ERCC standard validation. ERCC transcripts were added to the RNA sample during the construction of RNA sequencing (RNA-seq) library or targeted capture RNA sequencing (RNACap) library. The RPKM value of ERCC spike-in correlates linearly with ERCC spike-in concentration; transcripts of targeted genes enriched in RNACap were compared to RNA-seq.
Coverage of ERCC Transcripts
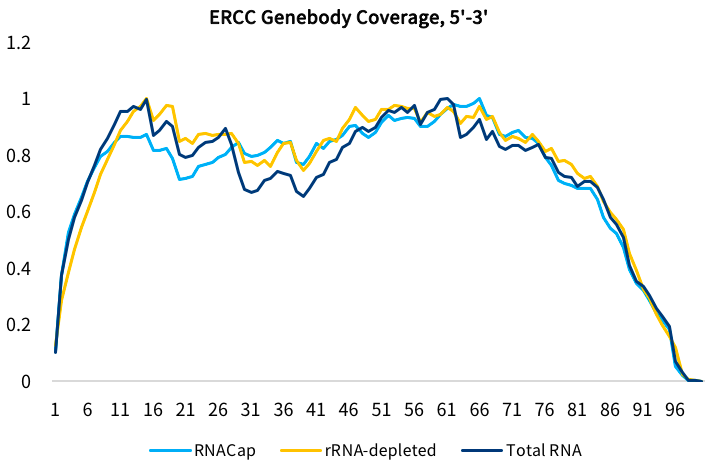
Fig 3. Influence of three RNA library construction methods on coverage of ERCC transcripts. Different methods achieving the same level of coverage with RNACap presents no coverage bias. For RNACap (blue), RNA library was constructed from total RNA for targeted capture sequencing; for rRNA-depleted (orange), RNA library was constructed for direct sequencing after ribosomal RNA (rRNA) been depleted from total RNA; for total RNA (dark blue), RNA library was constructed from total RNA for direct sequencing.
Performance Repeatability
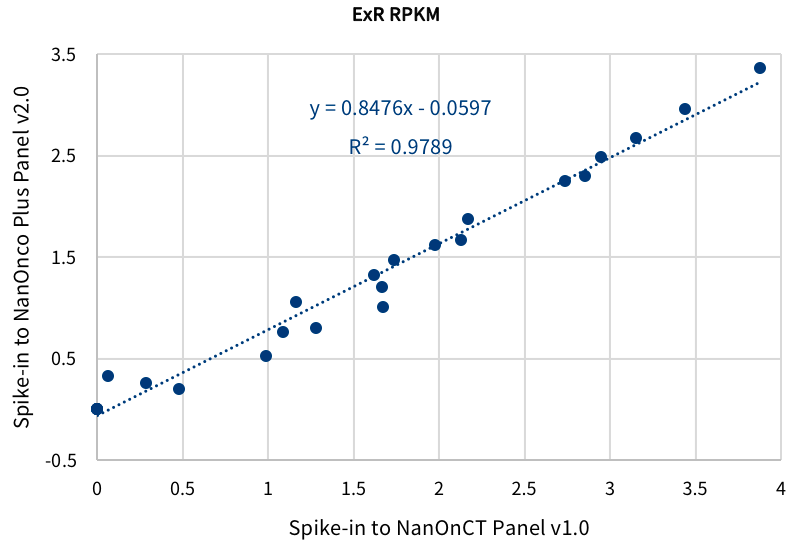
Fig 4. Performance repeatability of Ext-RNA Control Panel v1.0. The panel was spiked-in to Onco Plus Research Panel v2.0 and OnCT Research Panel v1.0 separately for targeted capture sequencing. Correlation analysis was introduced to analyze the repeatability of the log value of the RPKMs.
For research use only. Not for use in diagnostic procedures.
|
Catalog# |
Color Of Tube Cap |
Item |
Volume |
Package/Storage |
|
1001652 |
|
Ext-RNA Control Panel v1.0, 16 rxn |
70 μl×1 |
–20℃ |
|
1001651 |
|
Ext-RNA Control Panel v1.0, 96 rxn |
415 μl×1 |
–20℃ |
|
Product |
Catalog# |
|
Ext-RNA Control Panel v1.0, 16 rxn |
1001652 |
|
Ext-RNA Control Panel v1.0, 96 rxn |
1001651 |
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com