How is successful library preparation achieved for low quality and low input samples? Single-strand library preparation technology boosts success rate significantly!
View: 2521 / Time: 2024-09-03
01 Background
Library preparation is a critical step in all NGS workflows involving the processing of DNA/RNA samples into the appropriate insert size beginning with mechanical or enzymatic fragmentation, etc. Subsequently, the ends of the DNA are then repaired and primed for the ligation of specific sequencing adapters which are added to the 3’ and 5’ ends. This process results in the formation of an effective library that meets the requirements of the sequencing platform. Common adapter ligation methods for dsDNA library preparation include T-A ligation, transposase, blunt-end ligation, and cohesive-end ligation. However, conventional dsDNA library preparation methods typically yield unsatisfactory results when dealing with short fragment DNA, degraded DNA, ssDNA, or DNA samples with base damage, nicks, or gaps[1].
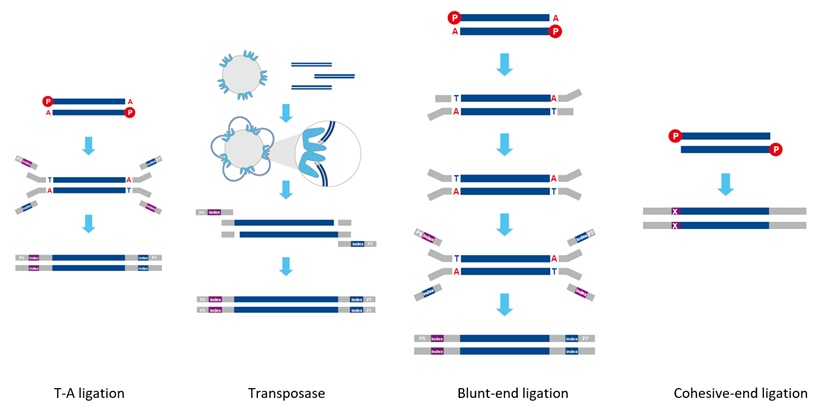
Figure 1. Common adapter ligation methods for dsDNA library preparation.
The ssDNA library preparation method has been proven to effectively preserve more information from shorter, degraded, and fragmented DNA[2]. It is considered a reliable approach for preparing libraries from low-quality, trace, or severely degraded samples. The strategy involves first denaturing the DNA sample at high temperatures to produce ssDNA. Then, adapters are stepwise ligated to both ends of the ssDNA to form a dsDNA structure, which is subsequently amplified by PCR to prepare the library.
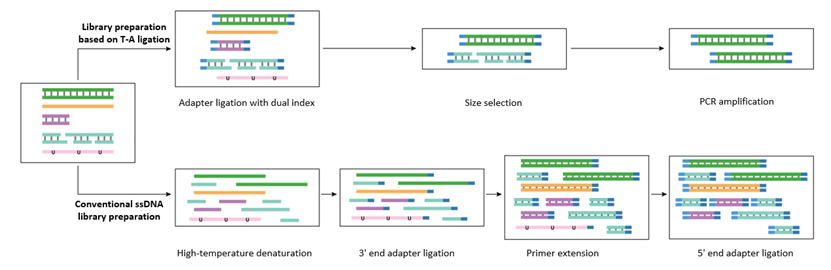
Figure 2. Handling capabilities of library preparation based on T-A ligation vs. conventional ssDNA ligation vary for different samples.
The single-strand library preparation technology eliminates the dependence on traditional T-A ligation from dsDNA. Consequently, it is no longer restricted by factors such as DNA end integrity, double-strand integrity, DNA input amount, or adapter/sample concentration ratio, thus maximizing DNA ligation efficiency and template utilization. It has significant enrichment advantages, particularly when processing samples such as ancient DNA, cfDNA/ctDNA, FFPE DNA, ChIP DNA, and bisulfite-converted products[3-5], thereby increasing the success rate of studies in ancient DNA research and cancer genomics/genetics based on liquid biopsy.
02 ssDNA Library Preparation Solution (for Illumina®)
2.1 Introduction
ssDNA Library Preparation Solution (for Illumina®) is developed based on the efficient single-stranded ligation principle, specifically designed for the Illumina® high-throughput sequencing platform. It is particularly suitable for library preparation from low-quality and ultralow-input samples, with initial input amounts ranging from 10 pg to 250 ng. It is applicable for whole-genome sequencing (WGS), whole-genome bisulfite sequencing (WGBS), and is also compatible with liquid-phase hybridization target capture sequencing. Optimized design ensures compatibility with cfDNA, gDNA, FFPE DNA samples, as well as bisulfite-converted DNA from cfDNA, gDNA and FFPE DNA samples.
2.2 Workflow
Wide Range of Sample Input: Adaptable to a broad range of sample input amounts (10 pg-250 ng), offering advantages over conventional dsDNA library preparation techniques.
Compatibility with Diverse Sample Types: Widely compatible with various sample types, including gDNA, cfDNA, FFPE DNA, and their bisulfite-converted products, ensuring versatility.
Comprehensive Compatibility with Multi-level FFPE Samples: Supporting for highly degraded low-quality FFPE samples, turning waste into treasure.
Efficient Library Preparation for Low-input/Trace Samples: Supports efficient library preparation from low-input/trace amounts of gDNA and cfDNA/ctDNA samples, ensuring stable and efficient library yield.
High-precision Methylation Library Preparation Solution: Providing library preparation solutions for bisulfite-converted products to meet the high-precision data requirements of methylation research.
03 Performance
3.1 Compatible with different input amounts of samples
3.2 Compatibility with diverse sample types
gDNA, FFPE DNA, and cfDNA, as well as their bisulfite-converted products, were used for single-strand library preparation with ssDNA Library Preparation Solution (for Illumina®). Satisfactory library yields were obtained for all samples (Fig. 5. A), with a concentrated distribution of library fragments. No noticeable adapter dimers were observed in the fragment distribution (Fig. 5. B).
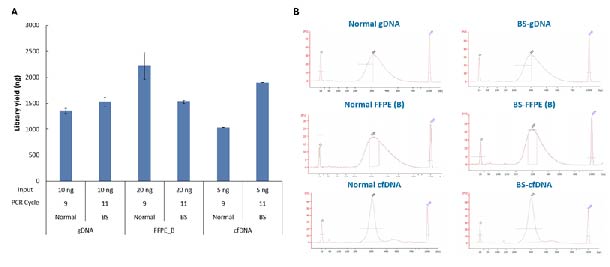
Figure 5. Library yield of ssDNA Library Preparation Solution for different types of samples. A. Library yield; B. Size distribution.
Note: FFPE samples are B-grade FFPE DNA. The grading standards by electrophoresis are as follows: FFPE B: one indistinct stripe with about 15 Kb in size, with medium diffusion.
3.3 Efficient library preparation for ultra-low-grade FFPE samples
Using ssDNA Library Preparation Solution (for Illumina®), single-strand library preparation was performed on B, C, and D-grade FFPE samples, and simultaneously compared with double-strand library preparation methods. The results showed that, at the same initial input amount, the library yield decreased as the quality grade of FFPE samples declines. However, for ultra-low-grade FFPE samples (FFPE_D) with lower initial inpout amounts, single-strand library preparation demonstrated a higher library yield compared to double-strand library preparation, showing significant advantages (Fig. 6. A). Furthermore, the advantages of single-strand library preparation for targeted sequencing become increasingly significant as the quality grade of the FFPE samples decreases. Specifically, single-strand library preparation exhibits higher mappability and on-target than double-strand library preparation (Fig. 6. B), as well as superior target covered (Fig. 6. C). Moreover, the average sequencing depth after deduplication is also higher (Fig. 6. D), especially for FFPE_D samples, where the deduplicated sequencing depth reaches four times that of double-strand library preparation.
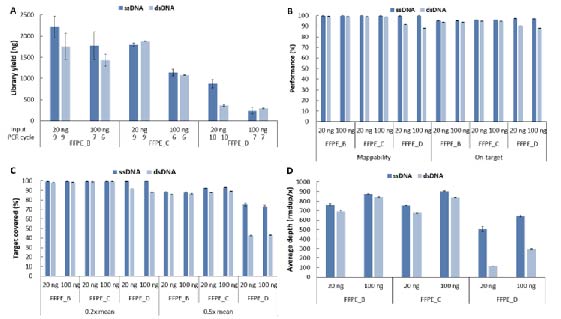
Figure 6. Comparison of different library preparation schemes on the library yield and capture performance of various grade FFPE DNA samples. A. Library yield; B. Mappability & On-target rate; C. Target covered; D. Average sequencing depth after deduplication. The capture was performed by using LungCancer Panel v1.0 coupled with NadPrep Hybrid Capture Reagents.
Note: The grading standards by electrophoresis are as follows: FFPE B: one indistinct stripe with about 15 kb in size, with medium diffusion. FFPE C: multiple stripes ranging from 200 bp to 2,500 bp, with severe diffusion. FFPE D: multiple stripes ranging from 250 bp to 1,000 bp, with severe diffusion.
3.4 Efficient library preparation for low-input samples
3.4.1 gDNA
Compared to conventional double-strand library preparation, single-strand library preparation shows significant advantages with low input amounts of samples. Single- and double-strand library preparations were performed on gDNA standards. In terms of library yield (Fig. 7. A), single-strand library preparation showed an absolute advantage at an input amount of 0.1 ng, with library yield being tens of times higher than that of double-strand library preparation. Additionally, target covered was significantly better with single-strand library preparation (Fig. 7. C). The average sequencing depth after deduplication was higher for single-strand library preparation compared to double-strand library preparation across all input amounts (Fig. 7. D). This helps to capture sample information more accurately, indirectly saving on sequencing data volume and thereby reducing detection costs.
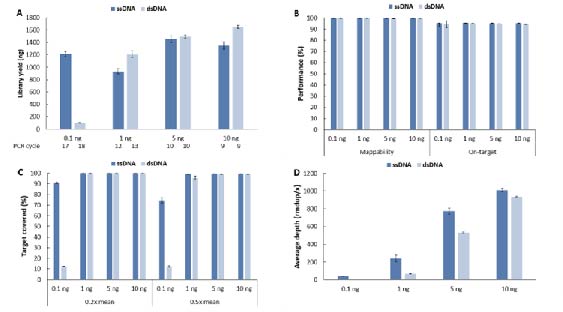
Figure 7. Comparison of different library preparation schemes on the library yield and capture performance of low-input gDNA samples. A. Library yield; B. Mappability & On-target rate; C. Target covered; D. Average sequencing depth after deduplication. The capture was performed by using LungCancer Panel v1.0 coupled with NadPrep Hybrid Capture Reagents. For each sample, 0.78 Gb of data was randomly selected for data analysis.
3.4.2 cfDNA
Similarly, we tested the library preparation for cfDNA at low input amounts, and the results were consistent with those for gDNA (Fig. 8). For library preparation with low input samples of less than 10 ng, the efficient ligation of single-strand library preparation compensates for the shortcomings of double-strand library preparation, making it a more suitable choice for library preparation.
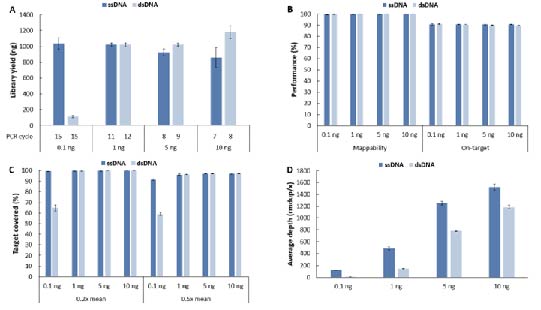
Figure 8. Comparison of Different Library Preparation Schemes on the library yield and capture performance of low-input cfDNA samples. A. Library yield; B. Mappability & On-target rate; C. Target covered; D. Average sequencing depth after deduplication. The capture was performed by using LungCancer Panel v1.0 coupled with NadPrep Hybrid Capture Reagents. For each sample, 0.81 Gb of data was randomly selected for data analysis.
ssDNA Library Preparation Solution (for Illumina®) not only has significant advantages with low-input and low-quality samples but also excels in methylation library preparation, providing a new approach for early cancer screening. For more test data, please follow Nanodigmbio. In our next issue, we will showcase more of the appeal of ssDNA library preparation.
References
[1] Gansauge M T, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA[J]. Nature protocols, 2013, 8(4): 737-748.
[2] Zhu J, Huang J, Zhang P, et al. Advantages of single-stranded DNA over double-stranded DNA library preparation for capturing cell-free tumor DNA in plasma[J]. Molecular diagnosis & therapy, 2020, 24: 95-101.
[3] Burnham P, Kim M S, Agbor-Enoh S, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma[J]. Scientific reports, 2016, 6(1): 27859.
[4] Stiller M, Sucker A, Griewank K, et al. Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA[J]. Oncotarget, 2016, 7(37): 59115.
[5] Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA[J]. PloS one, 2011, 6(9): e23418.
Library preparation is a critical step in all NGS workflows involving the processing of DNA/RNA samples into the appropriate insert size beginning with mechanical or enzymatic fragmentation, etc. Subsequently, the ends of the DNA are then repaired and primed for the ligation of specific sequencing adapters which are added to the 3’ and 5’ ends. This process results in the formation of an effective library that meets the requirements of the sequencing platform. Common adapter ligation methods for dsDNA library preparation include T-A ligation, transposase, blunt-end ligation, and cohesive-end ligation. However, conventional dsDNA library preparation methods typically yield unsatisfactory results when dealing with short fragment DNA, degraded DNA, ssDNA, or DNA samples with base damage, nicks, or gaps[1].

Figure 1. Common adapter ligation methods for dsDNA library preparation.
The ssDNA library preparation method has been proven to effectively preserve more information from shorter, degraded, and fragmented DNA[2]. It is considered a reliable approach for preparing libraries from low-quality, trace, or severely degraded samples. The strategy involves first denaturing the DNA sample at high temperatures to produce ssDNA. Then, adapters are stepwise ligated to both ends of the ssDNA to form a dsDNA structure, which is subsequently amplified by PCR to prepare the library.

Figure 2. Handling capabilities of library preparation based on T-A ligation vs. conventional ssDNA ligation vary for different samples.
The single-strand library preparation technology eliminates the dependence on traditional T-A ligation from dsDNA. Consequently, it is no longer restricted by factors such as DNA end integrity, double-strand integrity, DNA input amount, or adapter/sample concentration ratio, thus maximizing DNA ligation efficiency and template utilization. It has significant enrichment advantages, particularly when processing samples such as ancient DNA, cfDNA/ctDNA, FFPE DNA, ChIP DNA, and bisulfite-converted products[3-5], thereby increasing the success rate of studies in ancient DNA research and cancer genomics/genetics based on liquid biopsy.
02 ssDNA Library Preparation Solution (for Illumina®)
2.1 Introduction
ssDNA Library Preparation Solution (for Illumina®) is developed based on the efficient single-stranded ligation principle, specifically designed for the Illumina® high-throughput sequencing platform. It is particularly suitable for library preparation from low-quality and ultralow-input samples, with initial input amounts ranging from 10 pg to 250 ng. It is applicable for whole-genome sequencing (WGS), whole-genome bisulfite sequencing (WGBS), and is also compatible with liquid-phase hybridization target capture sequencing. Optimized design ensures compatibility with cfDNA, gDNA, FFPE DNA samples, as well as bisulfite-converted DNA from cfDNA, gDNA and FFPE DNA samples.
2.2 Workflow
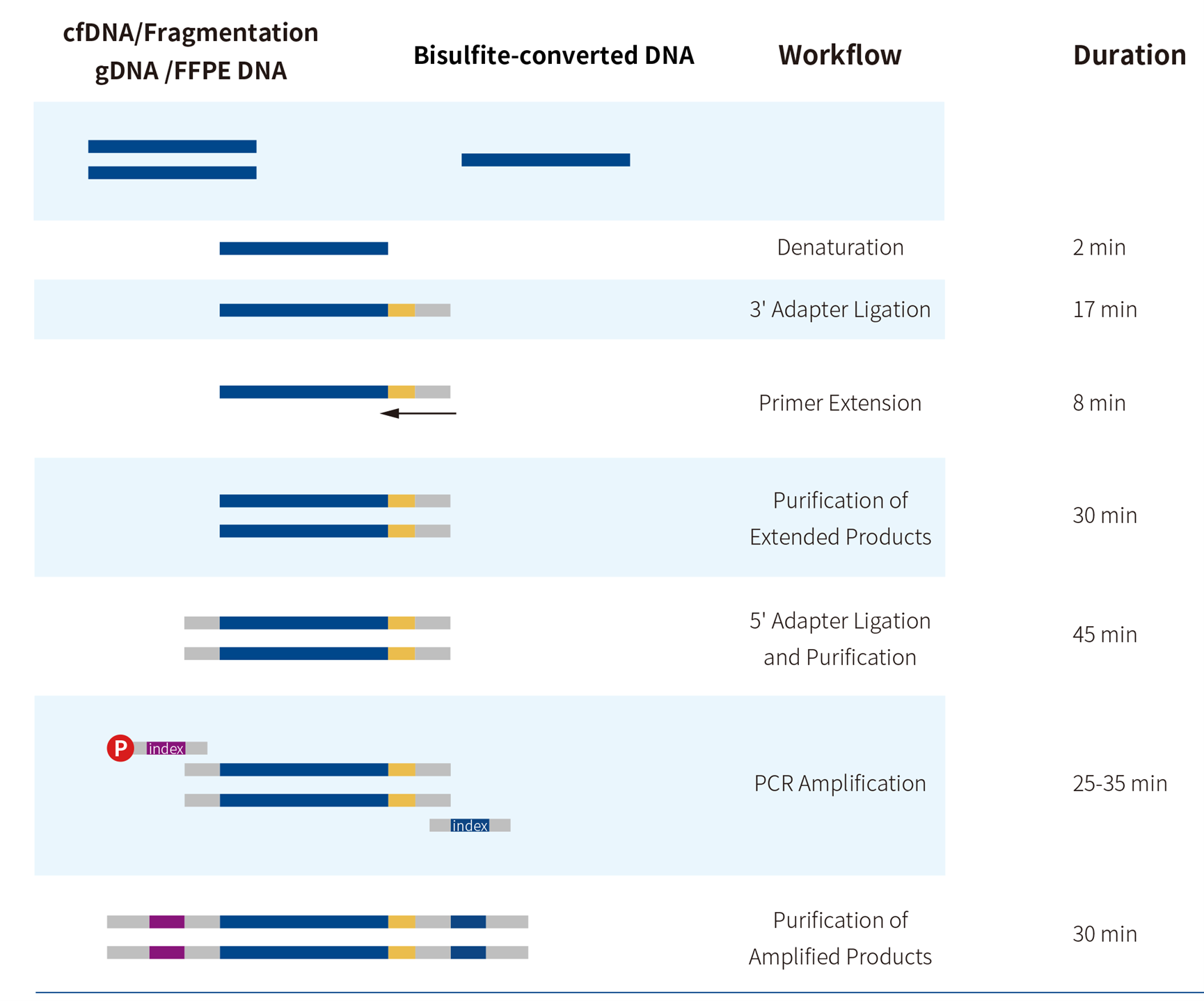
Figure 3. Workflow of ssDNA Library Preparation Solution.
2.3 FeaturesWide Range of Sample Input: Adaptable to a broad range of sample input amounts (10 pg-250 ng), offering advantages over conventional dsDNA library preparation techniques.
Compatibility with Diverse Sample Types: Widely compatible with various sample types, including gDNA, cfDNA, FFPE DNA, and their bisulfite-converted products, ensuring versatility.
Comprehensive Compatibility with Multi-level FFPE Samples: Supporting for highly degraded low-quality FFPE samples, turning waste into treasure.
Efficient Library Preparation for Low-input/Trace Samples: Supports efficient library preparation from low-input/trace amounts of gDNA and cfDNA/ctDNA samples, ensuring stable and efficient library yield.
High-precision Methylation Library Preparation Solution: Providing library preparation solutions for bisulfite-converted products to meet the high-precision data requirements of methylation research.
03 Performance
3.1 Compatible with different input amounts of samples
ssDNA Library Preparation Solution (for Illumina®) is suitable for library preparation from low-quality and ultralow-input samples, with initial input amounts ranging from 10 pg to 250 ng. Using the human genomic DNA standard (Promega, G1521) (Fig. 4. A) and its bisulfite-converted products (Fig. 4. B) for testing, with input amounts ranging from 10 pg to 250 ng, amplification was performed according to the recommended PCR cycle numbers. Satisfactory library yields were obtained for both types of gDNA across various input amount.
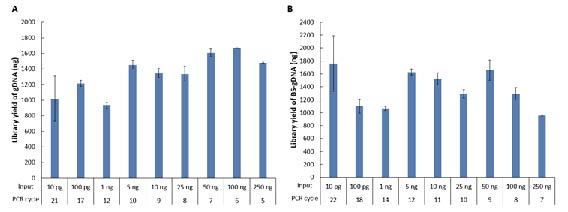
3.2 Compatibility with diverse sample types
gDNA, FFPE DNA, and cfDNA, as well as their bisulfite-converted products, were used for single-strand library preparation with ssDNA Library Preparation Solution (for Illumina®). Satisfactory library yields were obtained for all samples (Fig. 5. A), with a concentrated distribution of library fragments. No noticeable adapter dimers were observed in the fragment distribution (Fig. 5. B).

Figure 5. Library yield of ssDNA Library Preparation Solution for different types of samples. A. Library yield; B. Size distribution.
Note: FFPE samples are B-grade FFPE DNA. The grading standards by electrophoresis are as follows: FFPE B: one indistinct stripe with about 15 Kb in size, with medium diffusion.
3.3 Efficient library preparation for ultra-low-grade FFPE samples
Using ssDNA Library Preparation Solution (for Illumina®), single-strand library preparation was performed on B, C, and D-grade FFPE samples, and simultaneously compared with double-strand library preparation methods. The results showed that, at the same initial input amount, the library yield decreased as the quality grade of FFPE samples declines. However, for ultra-low-grade FFPE samples (FFPE_D) with lower initial inpout amounts, single-strand library preparation demonstrated a higher library yield compared to double-strand library preparation, showing significant advantages (Fig. 6. A). Furthermore, the advantages of single-strand library preparation for targeted sequencing become increasingly significant as the quality grade of the FFPE samples decreases. Specifically, single-strand library preparation exhibits higher mappability and on-target than double-strand library preparation (Fig. 6. B), as well as superior target covered (Fig. 6. C). Moreover, the average sequencing depth after deduplication is also higher (Fig. 6. D), especially for FFPE_D samples, where the deduplicated sequencing depth reaches four times that of double-strand library preparation.

Figure 6. Comparison of different library preparation schemes on the library yield and capture performance of various grade FFPE DNA samples. A. Library yield; B. Mappability & On-target rate; C. Target covered; D. Average sequencing depth after deduplication. The capture was performed by using LungCancer Panel v1.0 coupled with NadPrep Hybrid Capture Reagents.
Note: The grading standards by electrophoresis are as follows: FFPE B: one indistinct stripe with about 15 kb in size, with medium diffusion. FFPE C: multiple stripes ranging from 200 bp to 2,500 bp, with severe diffusion. FFPE D: multiple stripes ranging from 250 bp to 1,000 bp, with severe diffusion.
3.4 Efficient library preparation for low-input samples
3.4.1 gDNA
Compared to conventional double-strand library preparation, single-strand library preparation shows significant advantages with low input amounts of samples. Single- and double-strand library preparations were performed on gDNA standards. In terms of library yield (Fig. 7. A), single-strand library preparation showed an absolute advantage at an input amount of 0.1 ng, with library yield being tens of times higher than that of double-strand library preparation. Additionally, target covered was significantly better with single-strand library preparation (Fig. 7. C). The average sequencing depth after deduplication was higher for single-strand library preparation compared to double-strand library preparation across all input amounts (Fig. 7. D). This helps to capture sample information more accurately, indirectly saving on sequencing data volume and thereby reducing detection costs.

Figure 7. Comparison of different library preparation schemes on the library yield and capture performance of low-input gDNA samples. A. Library yield; B. Mappability & On-target rate; C. Target covered; D. Average sequencing depth after deduplication. The capture was performed by using LungCancer Panel v1.0 coupled with NadPrep Hybrid Capture Reagents. For each sample, 0.78 Gb of data was randomly selected for data analysis.
3.4.2 cfDNA
Similarly, we tested the library preparation for cfDNA at low input amounts, and the results were consistent with those for gDNA (Fig. 8). For library preparation with low input samples of less than 10 ng, the efficient ligation of single-strand library preparation compensates for the shortcomings of double-strand library preparation, making it a more suitable choice for library preparation.

Figure 8. Comparison of Different Library Preparation Schemes on the library yield and capture performance of low-input cfDNA samples. A. Library yield; B. Mappability & On-target rate; C. Target covered; D. Average sequencing depth after deduplication. The capture was performed by using LungCancer Panel v1.0 coupled with NadPrep Hybrid Capture Reagents. For each sample, 0.81 Gb of data was randomly selected for data analysis.
ssDNA Library Preparation Solution (for Illumina®) not only has significant advantages with low-input and low-quality samples but also excels in methylation library preparation, providing a new approach for early cancer screening. For more test data, please follow Nanodigmbio. In our next issue, we will showcase more of the appeal of ssDNA library preparation.
References
[1] Gansauge M T, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA[J]. Nature protocols, 2013, 8(4): 737-748.
[2] Zhu J, Huang J, Zhang P, et al. Advantages of single-stranded DNA over double-stranded DNA library preparation for capturing cell-free tumor DNA in plasma[J]. Molecular diagnosis & therapy, 2020, 24: 95-101.
[3] Burnham P, Kim M S, Agbor-Enoh S, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma[J]. Scientific reports, 2016, 6(1): 27859.
[4] Stiller M, Sucker A, Griewank K, et al. Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA[J]. Oncotarget, 2016, 7(37): 59115.
[5] Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA[J]. PloS one, 2011, 6(9): e23418.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)