How to Achieve Ultra-sensitive Synchronous Detection of Methylation and Mutation without Sequence Conversion?
View: 1390 / Time: 2024-08-28
01 How Can MSRE Replace BS Conversion to Achieve Methylation Detection?
Whole-genome bisulfite sequencing (WGBS) enables the detection of methylation status at the single-base resolution, allowing for the mapping of genome-wide methylation patterns. Numerous WGBS data indicate that the methylation status of CpG sites is spatially correlated, as 94% of CpG pairs within 1 Kb share the same methylation status. This phenomenon has been further confirmed in the recent study of germline cross-tissue CpG methylation characteristics (Figure 1.) [1].
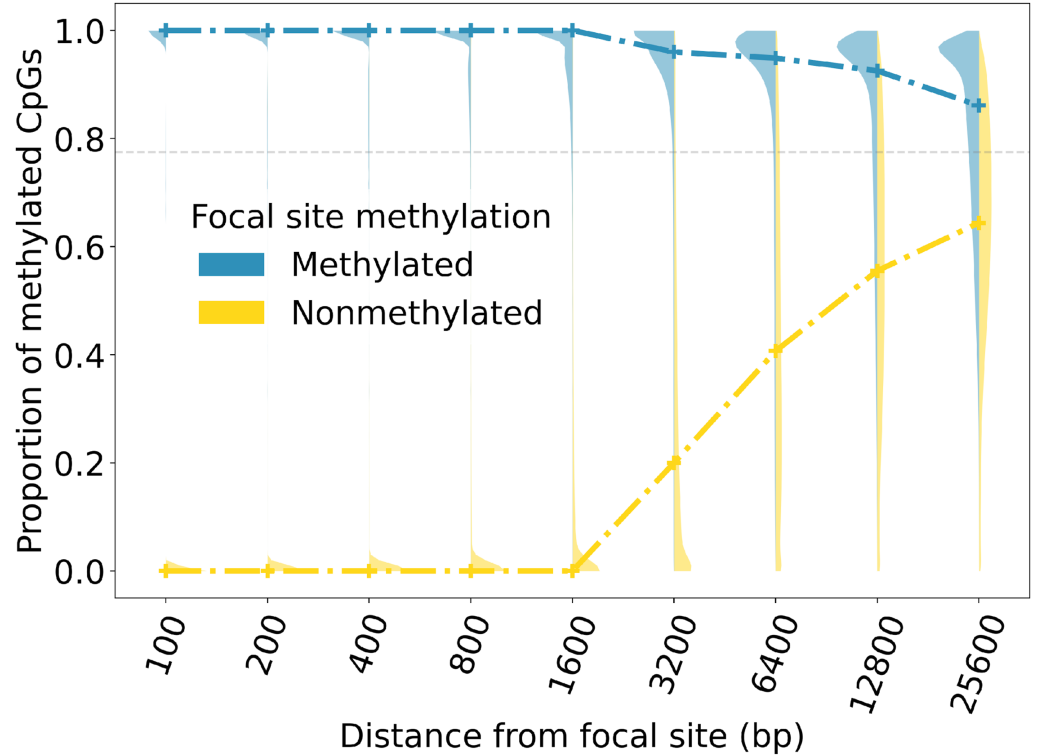
Figure 1. The methylation status of genomic CpG sites is spatially correlated[1]. The proportion of hypermethylated CpG sites around a focal site is related to its distance. Violin plots show the distribution of this proportion in varying neighborhood sizes; blue and yellow dashed lines mark the median values when the focal site is hyper- or hypo-methylated respectively
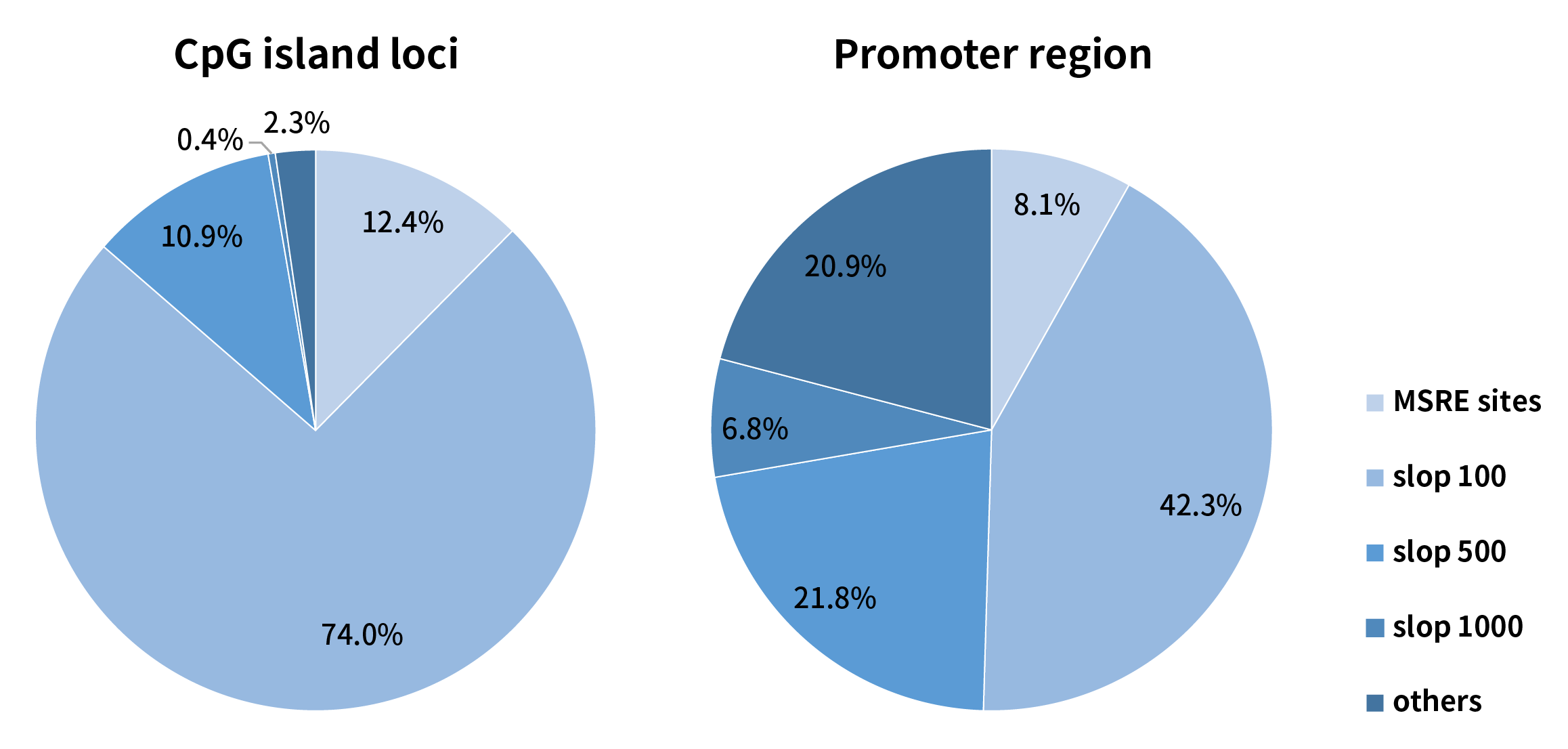
Methylation sensitive restriction endonucleases (MSRE) can only recognize the methylation status at specified sites. For example, the MSRE contained in Nanodigmbio’s μCaler™ DNA Full Screen System (hereinafter referred to as “Full Screen”, for details, please refer to the previous article: No Need for Conversion: Ultra-sensitive Solution for Synchronous Detection of Methylation and Mutation in a Single Reaction Coming Soon!) can only recognize and cover 12.4% of the CpG sites in the human genome. However, by sloping 500 bp on each side of the MSRE site, it is possible to cover 97.28% of the CpG sites. In the gene promoter region, MSRE can cover 8.1% of the CpG sites, and after sloping 500 bp on each side of the MSRE site, it can cover 72.29% of the CpG sites (Figure 2.). Therefore, considering the correlation of the methylation status, although the CpG sites directly recognized by MSRE are limited, they can still be used to analyze the methylation status within a certain regional range.
2.1 Significant Improvement in Effective Coverage Depth of Methylation Sites
Traditional BS treatment causes damage and degradation to DNA fragments during conversion. So, its limitations in application scenarios with a low amount of initial sample. However, MSRE treatment does not involve conversion and theoretically can retain more methylation site information. When using initial input of 10 ng, simulated samples with methylation levels of 0%, 2%, and 50% showed that the Full Screen group (treated with MSRE) achieved an average coverage depth of over 6 times that of the BS group for the same 13 CpG sites, approaching nearly 1,200x (Figure 3. A). When using initial inputs of 5 ng, 25 ng, and 50 ng, respectively, for simulated samples with a 5% methylation level, the average coverage depth of CpG sites in the Full Screen group was also several times higher than that of the BS group (Figure 3. B).
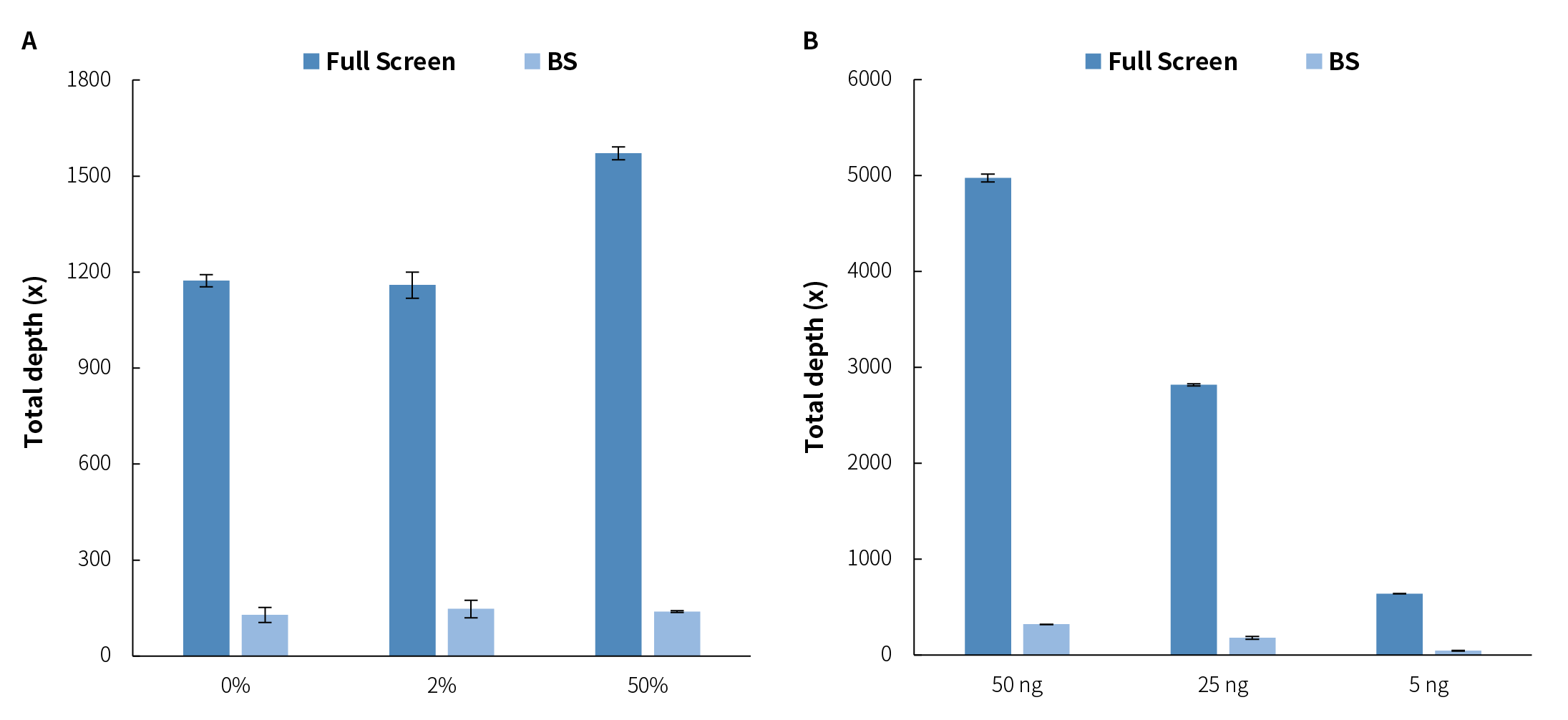 Figure 3. The μCaler™ DNA Full Screen System can increase the effective coverage depth of CpG sites several times that of the BS treatment. A. The average coverage depth of 13 CpG sites in simulated samples with different methylation levels at the initial inputs of 10 ng. B. The average coverage depth of 13 CpG sites in simulated samples with a methylation level of 5% at different initial input amounts. The samples were simulated DNA by using human 100% Methylated DNA standard (zymo, D5014-2) positive control and 0% Methylated DNA standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 2%, 5%, and 50%). The simulated samples were subjected to hybridization capture using the μCaler™ DNA Full Screen System Comprehensive Solution (Full Screen) and the μCaler™ targeted methylation capture solution (BS), respectively. The sequencing mode was Illumina® Novaseq 6000, PE150; 1 M reads pair were randomly selected for data analysis.
Figure 3. The μCaler™ DNA Full Screen System can increase the effective coverage depth of CpG sites several times that of the BS treatment. A. The average coverage depth of 13 CpG sites in simulated samples with different methylation levels at the initial inputs of 10 ng. B. The average coverage depth of 13 CpG sites in simulated samples with a methylation level of 5% at different initial input amounts. The samples were simulated DNA by using human 100% Methylated DNA standard (zymo, D5014-2) positive control and 0% Methylated DNA standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 2%, 5%, and 50%). The simulated samples were subjected to hybridization capture using the μCaler™ DNA Full Screen System Comprehensive Solution (Full Screen) and the μCaler™ targeted methylation capture solution (BS), respectively. The sequencing mode was Illumina® Novaseq 6000, PE150; 1 M reads pair were randomly selected for data analysis.
2.2 Precise Quantification of Methylation Level
The BS conversion method has high accuracy in DNA methylation detection and is considered the "gold standard". We obtained simulated samples with theoretical methylation levels of 0%, 2%, 5%, and 50% by mixing methylation DNA standards in different proportions to verify the consistency of the methylation status of the Full Screen group and the BS group. The results showed that the detected values of the methylation levels in four groups of simulated samples were consistent with the theoretical values, and the Full Screen group was also basically consistent with the BS group (Figure 4.). It is worth noting that the detection values of the Full Screen group were more stable and consistent, indicating that the detection results of the Full Screen can accurately reflect the true methylation status of the samples.
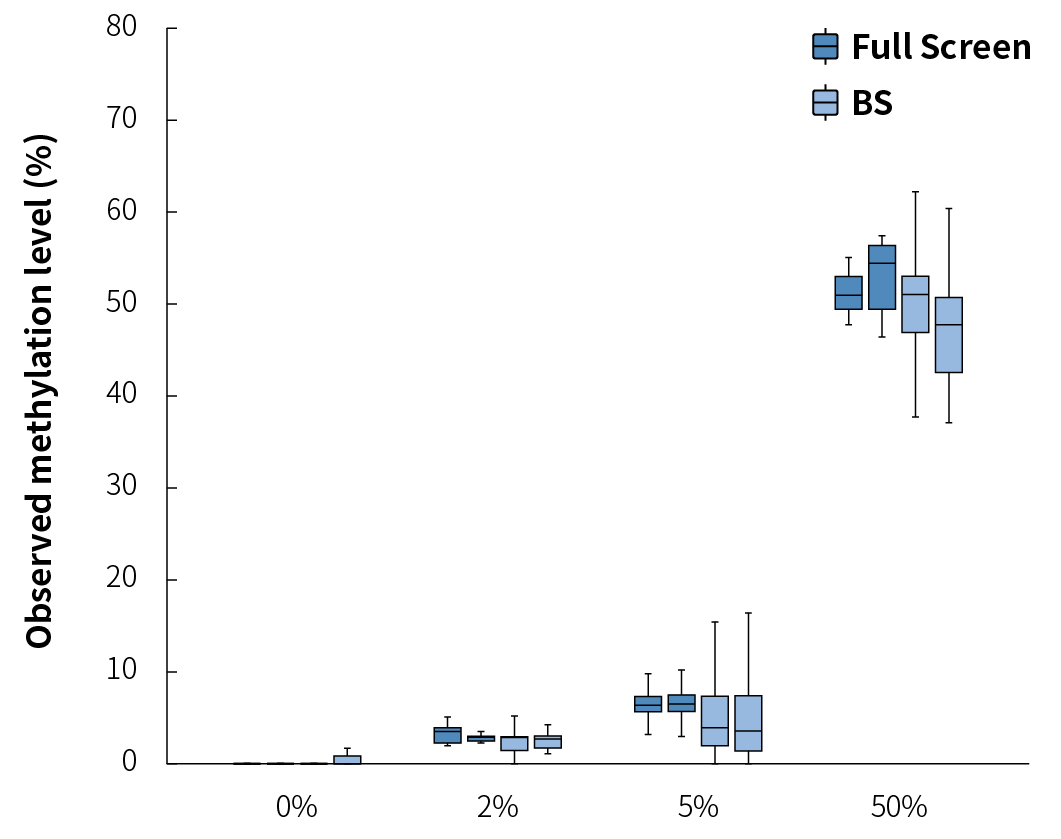
Figure 4. The detection results of the μCaler™ DNA Full Screen System for samples with different methylation levels are consistent with the theoretical values. The simulated samples were subjected to hybridization capture using the μCaler™ DNA Full Screen System Comprehensive Solution and the μCaler™ targeted methylation capture solution (BS), respectively. The sequencing mode was Illumina® Novaseq 6000, PE150; 1 M reads pair were randomly selected for data analysis.
2.3 Stable Cutting Efficiency of MSRE
Although the BS conversion efficiency is extremely high, it does not reach 100%. Unconverted cytosines during BS treatment may result in false positive signals, thereby affecting the accuracy of methylation detection. In experiments, exogenous unmethylated Lambda DNA is usually added as a reference to confirm whether the BS conversion rate meets expectations, with the standard being > 99%. Similar to BS conversion method, incomplete cleavage by MSRE can also lead to false positive signals.
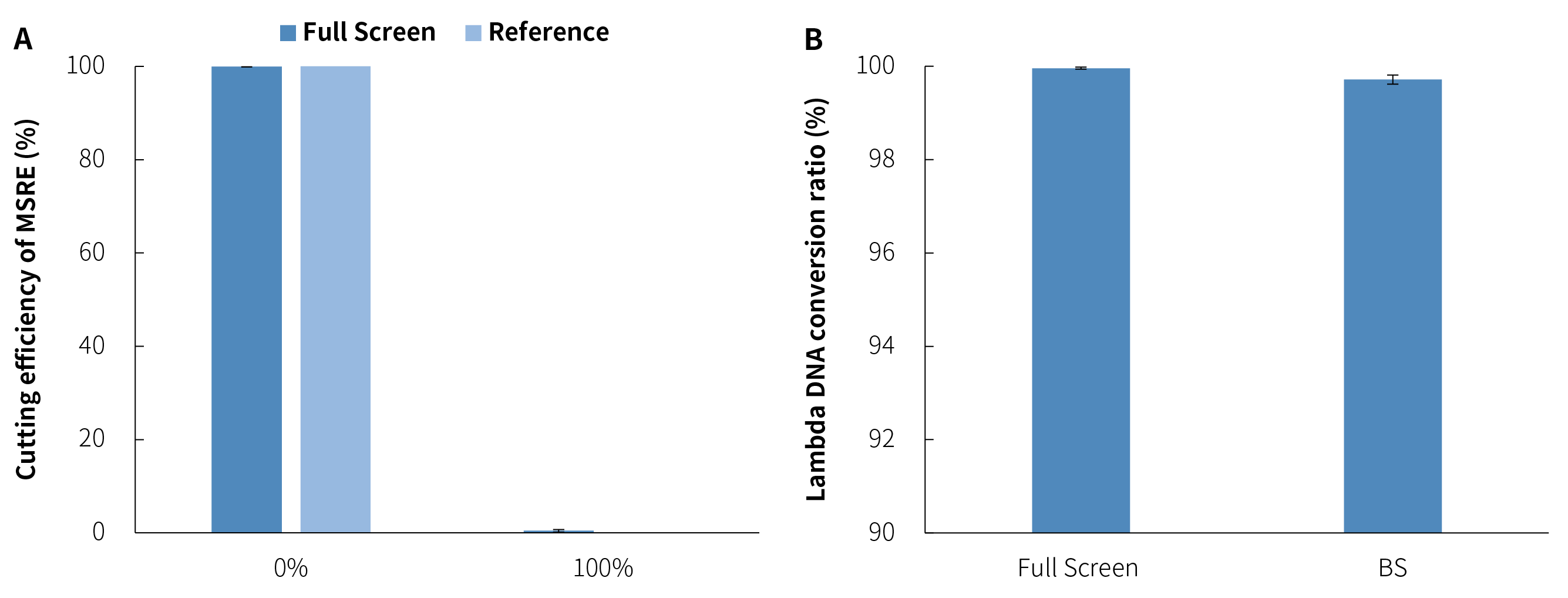
To evaluate the cutting efficiency of MSRE in the μCaler™ DNA Full Screen System, we also detected the cutting efficiency of unmethylated and fully methylated DNA standards by adding unmethylated Lambda DNA. As shown in Figure 5. A, the cutting efficiency of the Full Screen group for unmethylated sites reached 99.96%, while the cutting efficiency for fully methylated sites was only 0.42%, which was almost impossible to cut. This indicates that the MSRE in the μCaler™ DNA Full Screen System is sensitive to methylation sites while being highly efficient in cutting unmethylated sites.
We used the cutting efficiency of unmethylated Lambda DNA in the Full Screen group as an analogy to the "conversion rate" and compared it to the conversion rate of the same sites using the traditional BS conversion method. As shown in Figure 5. B, the MSRE conversion ratesfor Lambda DNA were highly stable across different batches, all exceeding 99.5%, slightly better than those achieved by the BS conversion method.
In healthy individuals, the median concentration of cfDNA has been estimated at only 1.2 - 2.4 ng/mL of plasma, while although the concentration could be increased significantly in cancer patients, the median value is still only 2.5 ng/mL[2]. How to achieve full use of these precious cfDNA samples to detect more biomarkers is one of the key technical challenges in early cancer screening and MRD monitoring. Taking methylation and mutation as examples of major biomarkers, traditional detection method (Classical) requires splitting the limited samples in two for separate tests, as DNA methylation detection relies on BS conversion. However, even with the deep optimizations without the change of principle, “”this approach still limited by a maximum sample utilization rate of 50%.
The μCaler™ DNA Full Screen System overcomes this bottleneck by allowing the synchronous detection of methylation and mutation in a single reaction, undoubtedly greatly improving the utilization efficiency of low initial amount samples, especially cfDNA (Figure 6.).
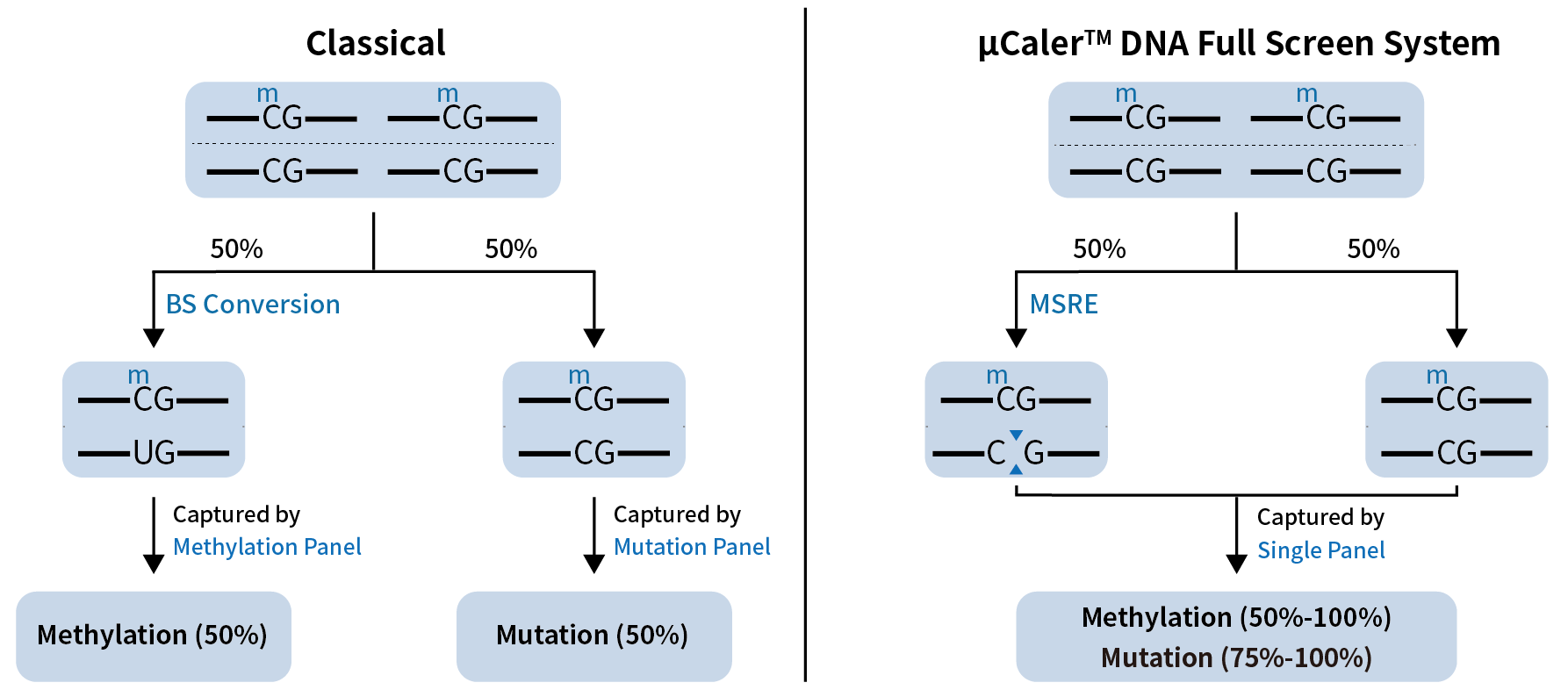 Figure 6. The principle of synchronous detection of methylation and mutation in a single reaction by μCaler™ DNA Full Screen System.
Figure 6. The principle of synchronous detection of methylation and mutation in a single reaction by μCaler™ DNA Full Screen System.
3.1 Full Screen Significantly Improves the Sensitivity of Low-Frequency Mutation Detection
When performing synchronous detection of methylation and mutation, the Full Screen can utilize all the initial samples for mutation analysis, thus providing more available templates and enhancing the detection sensitivity. Through DNA standards and gradient dilutions (1:10 and 1:100), we constructed simulated samples with methylation levels of approximately 40%, 4%, and 0.4% (31 CpG sites) respectively, and corresponding average mutation frequencies of 2%, 0.2%, and 0.02% (3 SNV mutation sites). For synchronous detection, the Full Screen group used the entire 80 ng of initial input samples for both mutation and methylation analysis; while the Classical group was carried out in a way that 50% (20 ng) was used for mutation analysis and 50% was used for methylation analysis. As shown in Figure 7., benefiting from more input amounts, the Full Screen group achieved approximately double the average effective depth compared to the Classical group, and detected more supporting mutation reads at the three different mutation frequency gradients (142 vs. 76; 15 vs. 10; 1 vs. 0).
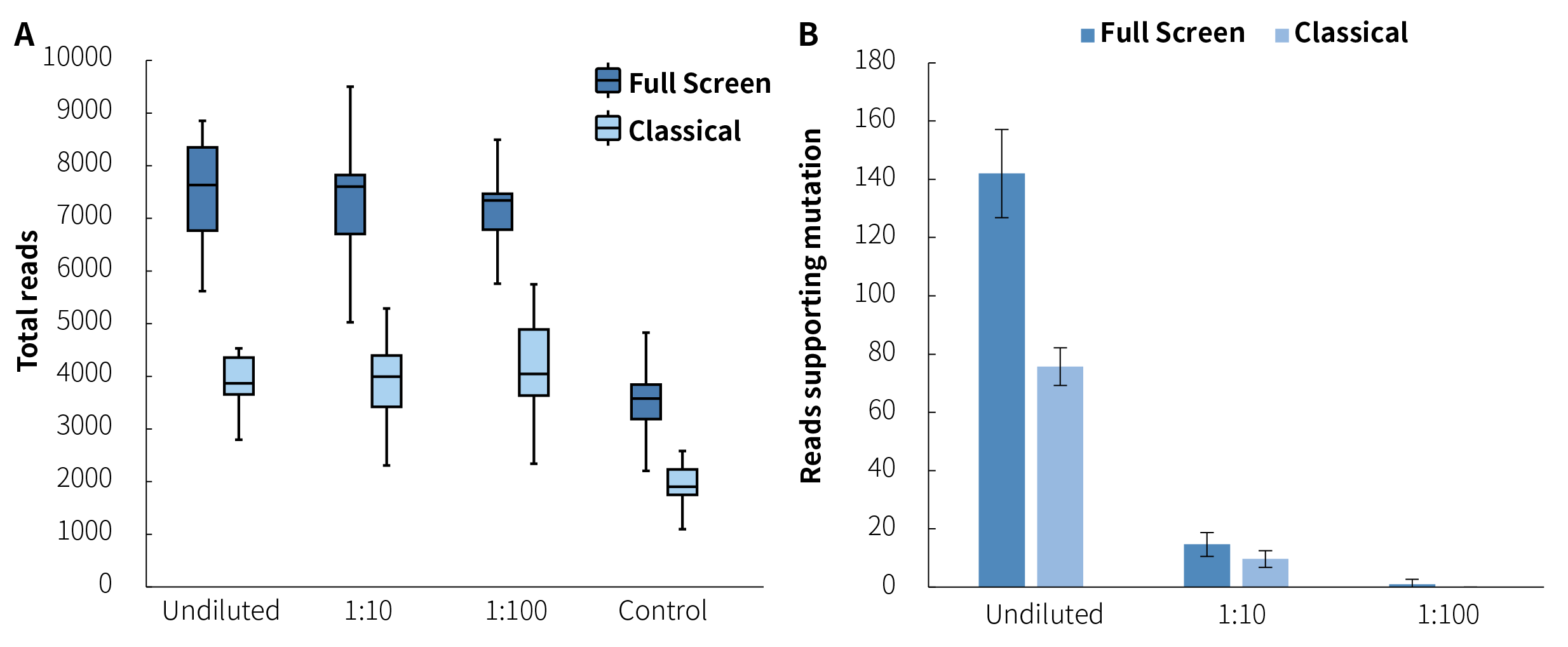 Figure 7. μCaler™ DNA Full Screen System can achieve double the average effective depth of mutations compared to the Classical method. A. The average coverage depth of mutation sites; B. The average count of reads supporting mutation [pre-library containing UMI, analyzed by Duplex Consensus Sequences (DCS211)]. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).
Figure 7. μCaler™ DNA Full Screen System can achieve double the average effective depth of mutations compared to the Classical method. A. The average coverage depth of mutation sites; B. The average count of reads supporting mutation [pre-library containing UMI, analyzed by Duplex Consensus Sequences (DCS211)]. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).
3.2 Examples of Synchronous Detection Results
In addition to the conventional mutation detections such as SNPs, fusions and deletions, μCaler™ DNA Full Screen System allows for simultaneously methylation detection. Figure 8. shows the synchronous detection results in the above-mentioned gradient mutations and methylation standards. The detection results of different mutation types and frequencies are basically consistent with the theoretical values (Figure 8. A). Meanwhile, the detection values of different methylation sites are in line with expectations, with clear observed gradients between different groups (Figure 8. B).
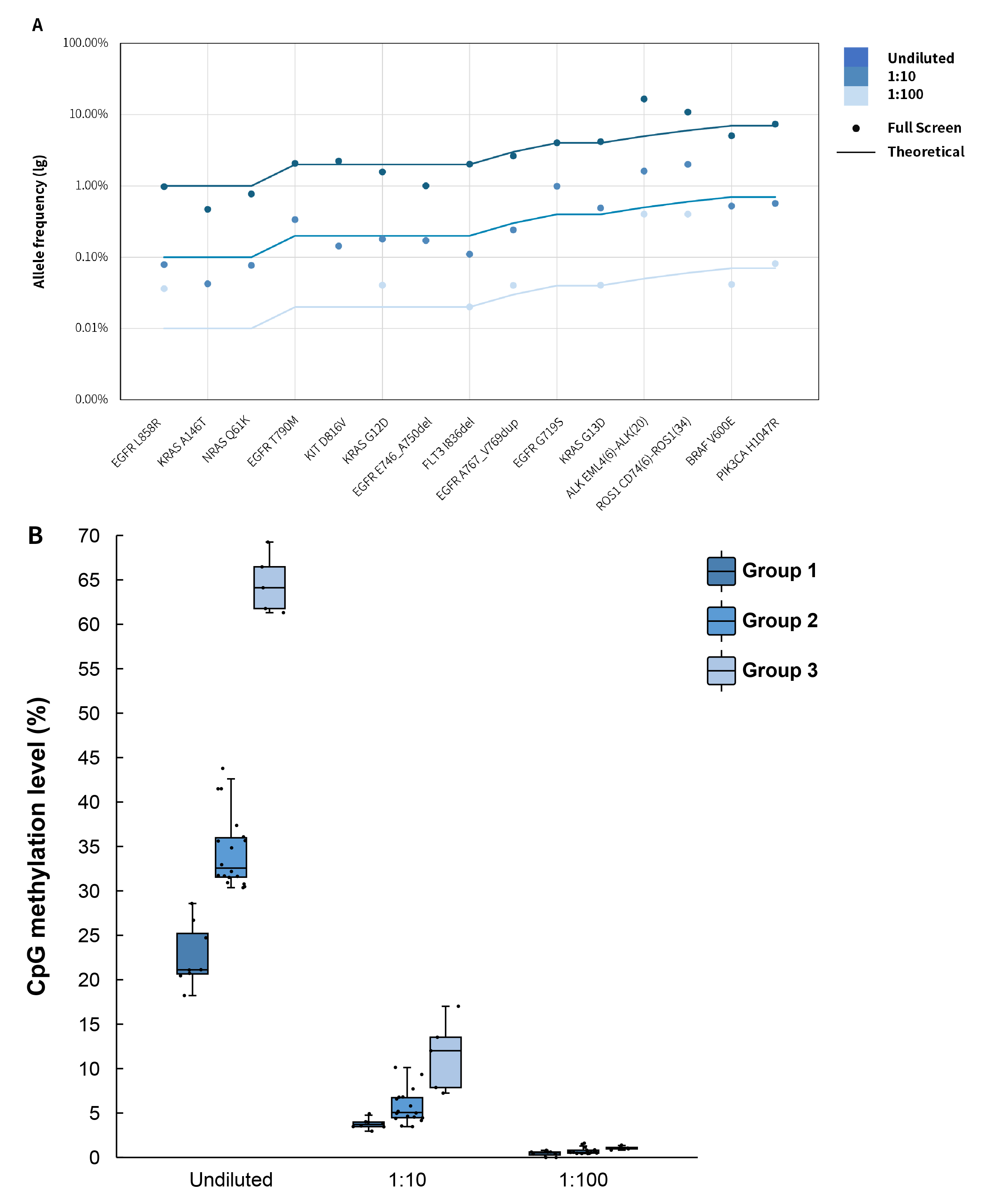 Figure 8. The performance of μCaler™ DNA Full Screen System applied to synchronous detection of methylation and mutation. A. The mutation frequencies at different sites were consistent with the nominal frequencies of the DNA standards; B. The average methylation level of different simulated samples across the three groups with different gradients in methylation level. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).
Figure 8. The performance of μCaler™ DNA Full Screen System applied to synchronous detection of methylation and mutation. A. The mutation frequencies at different sites were consistent with the nominal frequencies of the DNA standards; B. The average methylation level of different simulated samples across the three groups with different gradients in methylation level. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).
μCaler™ DNA Full Screen System is developed based on Nanodigmbio's exclusive patented ultra-sensitive μCaler™ Hybrid Capture technology. It operates with just one initial sample, one hybridization reaction, and one set of capture probes to achieve ultra-sensitive synchronous detection of methylation and mutation, with no need for sequence conversion. This provides powerful technical support for the precise diagnosis, treatment and prognosis assessment of tumors. This solution includes comprehensive services from scheme design and fully self-developed reagents to wet-lab support and bioinformatics analysis, making it user-friendly and easy to master.
Reference
[1] Yichen Si, Hyun Min Kang, Sebastian Zollner. Germline CpG methylation signatures in the human population inferred from genetic polymorphism[J]. bioRxiv, 2024.
[2] van der Pol Y, Mouliere F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA [J]. Cancer Cell, 2019, 36(4):350-368.
Whole-genome bisulfite sequencing (WGBS) enables the detection of methylation status at the single-base resolution, allowing for the mapping of genome-wide methylation patterns. Numerous WGBS data indicate that the methylation status of CpG sites is spatially correlated, as 94% of CpG pairs within 1 Kb share the same methylation status. This phenomenon has been further confirmed in the recent study of germline cross-tissue CpG methylation characteristics (Figure 1.) [1].

Figure 1. The methylation status of genomic CpG sites is spatially correlated[1]. The proportion of hypermethylated CpG sites around a focal site is related to its distance. Violin plots show the distribution of this proportion in varying neighborhood sizes; blue and yellow dashed lines mark the median values when the focal site is hyper- or hypo-methylated respectively
Methylation sensitive restriction endonucleases (MSRE) can only recognize the methylation status at specified sites. For example, the MSRE contained in Nanodigmbio’s μCaler™ DNA Full Screen System (hereinafter referred to as “Full Screen”, for details, please refer to the previous article: No Need for Conversion: Ultra-sensitive Solution for Synchronous Detection of Methylation and Mutation in a Single Reaction Coming Soon!) can only recognize and cover 12.4% of the CpG sites in the human genome. However, by sloping 500 bp on each side of the MSRE site, it is possible to cover 97.28% of the CpG sites. In the gene promoter region, MSRE can cover 8.1% of the CpG sites, and after sloping 500 bp on each side of the MSRE site, it can cover 72.29% of the CpG sites (Figure 2.). Therefore, considering the correlation of the methylation status, although the CpG sites directly recognized by MSRE are limited, they can still be used to analyze the methylation status within a certain regional range.

Figure 2. Distribution of CpG sites recognized by MSRE in the μCaler™ DNA Full Screen System within the CpG islands and promoters regions in human genome.
2.1 Significant Improvement in Effective Coverage Depth of Methylation Sites
Traditional BS treatment causes damage and degradation to DNA fragments during conversion. So, its limitations in application scenarios with a low amount of initial sample. However, MSRE treatment does not involve conversion and theoretically can retain more methylation site information. When using initial input of 10 ng, simulated samples with methylation levels of 0%, 2%, and 50% showed that the Full Screen group (treated with MSRE) achieved an average coverage depth of over 6 times that of the BS group for the same 13 CpG sites, approaching nearly 1,200x (Figure 3. A). When using initial inputs of 5 ng, 25 ng, and 50 ng, respectively, for simulated samples with a 5% methylation level, the average coverage depth of CpG sites in the Full Screen group was also several times higher than that of the BS group (Figure 3. B).
 Figure 3. The μCaler™ DNA Full Screen System can increase the effective coverage depth of CpG sites several times that of the BS treatment. A. The average coverage depth of 13 CpG sites in simulated samples with different methylation levels at the initial inputs of 10 ng. B. The average coverage depth of 13 CpG sites in simulated samples with a methylation level of 5% at different initial input amounts. The samples were simulated DNA by using human 100% Methylated DNA standard (zymo, D5014-2) positive control and 0% Methylated DNA standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 2%, 5%, and 50%). The simulated samples were subjected to hybridization capture using the μCaler™ DNA Full Screen System Comprehensive Solution (Full Screen) and the μCaler™ targeted methylation capture solution (BS), respectively. The sequencing mode was Illumina® Novaseq 6000, PE150; 1 M reads pair were randomly selected for data analysis.
Figure 3. The μCaler™ DNA Full Screen System can increase the effective coverage depth of CpG sites several times that of the BS treatment. A. The average coverage depth of 13 CpG sites in simulated samples with different methylation levels at the initial inputs of 10 ng. B. The average coverage depth of 13 CpG sites in simulated samples with a methylation level of 5% at different initial input amounts. The samples were simulated DNA by using human 100% Methylated DNA standard (zymo, D5014-2) positive control and 0% Methylated DNA standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 2%, 5%, and 50%). The simulated samples were subjected to hybridization capture using the μCaler™ DNA Full Screen System Comprehensive Solution (Full Screen) and the μCaler™ targeted methylation capture solution (BS), respectively. The sequencing mode was Illumina® Novaseq 6000, PE150; 1 M reads pair were randomly selected for data analysis.2.2 Precise Quantification of Methylation Level
The BS conversion method has high accuracy in DNA methylation detection and is considered the "gold standard". We obtained simulated samples with theoretical methylation levels of 0%, 2%, 5%, and 50% by mixing methylation DNA standards in different proportions to verify the consistency of the methylation status of the Full Screen group and the BS group. The results showed that the detected values of the methylation levels in four groups of simulated samples were consistent with the theoretical values, and the Full Screen group was also basically consistent with the BS group (Figure 4.). It is worth noting that the detection values of the Full Screen group were more stable and consistent, indicating that the detection results of the Full Screen can accurately reflect the true methylation status of the samples.

Figure 4. The detection results of the μCaler™ DNA Full Screen System for samples with different methylation levels are consistent with the theoretical values. The simulated samples were subjected to hybridization capture using the μCaler™ DNA Full Screen System Comprehensive Solution and the μCaler™ targeted methylation capture solution (BS), respectively. The sequencing mode was Illumina® Novaseq 6000, PE150; 1 M reads pair were randomly selected for data analysis.
2.3 Stable Cutting Efficiency of MSRE
Although the BS conversion efficiency is extremely high, it does not reach 100%. Unconverted cytosines during BS treatment may result in false positive signals, thereby affecting the accuracy of methylation detection. In experiments, exogenous unmethylated Lambda DNA is usually added as a reference to confirm whether the BS conversion rate meets expectations, with the standard being > 99%. Similar to BS conversion method, incomplete cleavage by MSRE can also lead to false positive signals.
To evaluate the cutting efficiency of MSRE in the μCaler™ DNA Full Screen System, we also detected the cutting efficiency of unmethylated and fully methylated DNA standards by adding unmethylated Lambda DNA. As shown in Figure 5. A, the cutting efficiency of the Full Screen group for unmethylated sites reached 99.96%, while the cutting efficiency for fully methylated sites was only 0.42%, which was almost impossible to cut. This indicates that the MSRE in the μCaler™ DNA Full Screen System is sensitive to methylation sites while being highly efficient in cutting unmethylated sites.
We used the cutting efficiency of unmethylated Lambda DNA in the Full Screen group as an analogy to the "conversion rate" and compared it to the conversion rate of the same sites using the traditional BS conversion method. As shown in Figure 5. B, the MSRE conversion ratesfor Lambda DNA were highly stable across different batches, all exceeding 99.5%, slightly better than those achieved by the BS conversion method.

Figure 5. The μCaler™ DNA Full Screen System can stably and efficiently cut the unmethylated sites. A. The cutting efficiency of unmethylated and fully methylated MSRE sites; B. The conversion rate of Lambda DNA.
In healthy individuals, the median concentration of cfDNA has been estimated at only 1.2 - 2.4 ng/mL of plasma, while although the concentration could be increased significantly in cancer patients, the median value is still only 2.5 ng/mL[2]. How to achieve full use of these precious cfDNA samples to detect more biomarkers is one of the key technical challenges in early cancer screening and MRD monitoring. Taking methylation and mutation as examples of major biomarkers, traditional detection method (Classical) requires splitting the limited samples in two for separate tests, as DNA methylation detection relies on BS conversion. However, even with the deep optimizations without the change of principle, “”this approach still limited by a maximum sample utilization rate of 50%.
The μCaler™ DNA Full Screen System overcomes this bottleneck by allowing the synchronous detection of methylation and mutation in a single reaction, undoubtedly greatly improving the utilization efficiency of low initial amount samples, especially cfDNA (Figure 6.).
 Figure 6. The principle of synchronous detection of methylation and mutation in a single reaction by μCaler™ DNA Full Screen System.
Figure 6. The principle of synchronous detection of methylation and mutation in a single reaction by μCaler™ DNA Full Screen System.3.1 Full Screen Significantly Improves the Sensitivity of Low-Frequency Mutation Detection
When performing synchronous detection of methylation and mutation, the Full Screen can utilize all the initial samples for mutation analysis, thus providing more available templates and enhancing the detection sensitivity. Through DNA standards and gradient dilutions (1:10 and 1:100), we constructed simulated samples with methylation levels of approximately 40%, 4%, and 0.4% (31 CpG sites) respectively, and corresponding average mutation frequencies of 2%, 0.2%, and 0.02% (3 SNV mutation sites). For synchronous detection, the Full Screen group used the entire 80 ng of initial input samples for both mutation and methylation analysis; while the Classical group was carried out in a way that 50% (20 ng) was used for mutation analysis and 50% was used for methylation analysis. As shown in Figure 7., benefiting from more input amounts, the Full Screen group achieved approximately double the average effective depth compared to the Classical group, and detected more supporting mutation reads at the three different mutation frequency gradients (142 vs. 76; 15 vs. 10; 1 vs. 0).
 Figure 7. μCaler™ DNA Full Screen System can achieve double the average effective depth of mutations compared to the Classical method. A. The average coverage depth of mutation sites; B. The average count of reads supporting mutation [pre-library containing UMI, analyzed by Duplex Consensus Sequences (DCS211)]. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).
Figure 7. μCaler™ DNA Full Screen System can achieve double the average effective depth of mutations compared to the Classical method. A. The average coverage depth of mutation sites; B. The average count of reads supporting mutation [pre-library containing UMI, analyzed by Duplex Consensus Sequences (DCS211)]. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).3.2 Examples of Synchronous Detection Results
In addition to the conventional mutation detections such as SNPs, fusions and deletions, μCaler™ DNA Full Screen System allows for simultaneously methylation detection. Figure 8. shows the synchronous detection results in the above-mentioned gradient mutations and methylation standards. The detection results of different mutation types and frequencies are basically consistent with the theoretical values (Figure 8. A). Meanwhile, the detection values of different methylation sites are in line with expectations, with clear observed gradients between different groups (Figure 8. B).
 Figure 8. The performance of μCaler™ DNA Full Screen System applied to synchronous detection of methylation and mutation. A. The mutation frequencies at different sites were consistent with the nominal frequencies of the DNA standards; B. The average methylation level of different simulated samples across the three groups with different gradients in methylation level. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).
Figure 8. The performance of μCaler™ DNA Full Screen System applied to synchronous detection of methylation and mutation. A. The mutation frequencies at different sites were consistent with the nominal frequencies of the DNA standards; B. The average methylation level of different simulated samples across the three groups with different gradients in methylation level. Hybrid capture was used μCaler™ Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites).

μCaler™ DNA Full Screen System is developed based on Nanodigmbio's exclusive patented ultra-sensitive μCaler™ Hybrid Capture technology. It operates with just one initial sample, one hybridization reaction, and one set of capture probes to achieve ultra-sensitive synchronous detection of methylation and mutation, with no need for sequence conversion. This provides powerful technical support for the precise diagnosis, treatment and prognosis assessment of tumors. This solution includes comprehensive services from scheme design and fully self-developed reagents to wet-lab support and bioinformatics analysis, making it user-friendly and easy to master.
In the subsequent chapters, we will present application cases in clinical samples. Stay tuned!
Welcome to contact sales@njnad.com or local sales for more information.
Reference
[1] Yichen Si, Hyun Min Kang, Sebastian Zollner. Germline CpG methylation signatures in the human population inferred from genetic polymorphism[J]. bioRxiv, 2024.
[2] van der Pol Y, Mouliere F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA [J]. Cancer Cell, 2019, 36(4):350-368.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)