New Arrival | HLAtyping Panel v2.0 Supports high precision typing to the fourth field level
Human leukocyte antigen (HLA), the major histocompatibility complex (MHC) of humans, is located on the short arm of chromosome 6 and is an important part of the human immune system. Different complexes bind to various exogenous or endogenous antigenic fragments through their binding grooves, which further interact with different T cells to activate downstream immune responses. Therefore, the functions undertaken by HLA place demands on its own diversity: HLA genes are among the most polymorphic region of human genome. The probability of two unrelated people having the same HLA gene combination is extremely low, and the immune system also distinguishes itself from foreign cells through the HLA on the cell surface. Foreign cells with different HLAs will be recognized as "not me" and will cause rejection. For a long time, HLA typing has been widely carried out in the field of organ transplantation, forming a complete set of systems and norms. HLA is also used in forensics as a molecular identifier that differentiates individuals. In addition, the rapid development of tumor immunotherapy research is also inseparable from the accurate typing of HLA genes.
The current HLA nomenclature has been implemented since 2010. There are four fields s after the gene name, which distinguish allele group, specific HLA protein, synonymous polymorphisms, and sequence polymorphisms in non-coding regions in turn. The suffix letters are annotate the expression level of this type. From the version 3.0 in April 2010 to the current version 3.50, the number of alleles included in the IPD-IMGT/HLA database has increased from more than 4,000 to more than 30,000, which is closely related to the widespread application of NGS in HLA typing in recent years.
Commonly NGS strategy is to amplify HLA gene fragments by long-range PCR, then fragment the amplified products to prepare the library for sequencing and sequence analysis. Obtaining the full-length sequence of the HLA gene is much less difficult than Sanger sequencing, and large number of new types of non-coding sequences with differences in the fourth field level have also been accumulated in the database.
Nanodigmbio launched the HLAtyping Panel v1.0 in 2019 to make targeted sequencing with hybridization capture for coding region of the HLA gene, which is design coding region probes based on the reference genome sequence, then investigate the sequence differences of different types to supplement polymorphism probes. It solves the problem that the polymorphism of HLA sequence exceeds the error tolerance of a single probe, and realizes the effective capture of all types of coding regions.
Combined with the background of the continuous update of database and the continuous enrichment of full-length sequences, we have launched HLAtyping Panel v2.0, which is refers to the latest database (Version 3.50), and targets 11 HLA genes (Class I: HLA-A, B, C; Class II: HLA-DPA1, DPB1, DQA1, DAQB1, DRB1/3/4/5). The probe covers the full-length genome sequence of all alleles and supports high precision typing up to the fourth field level.
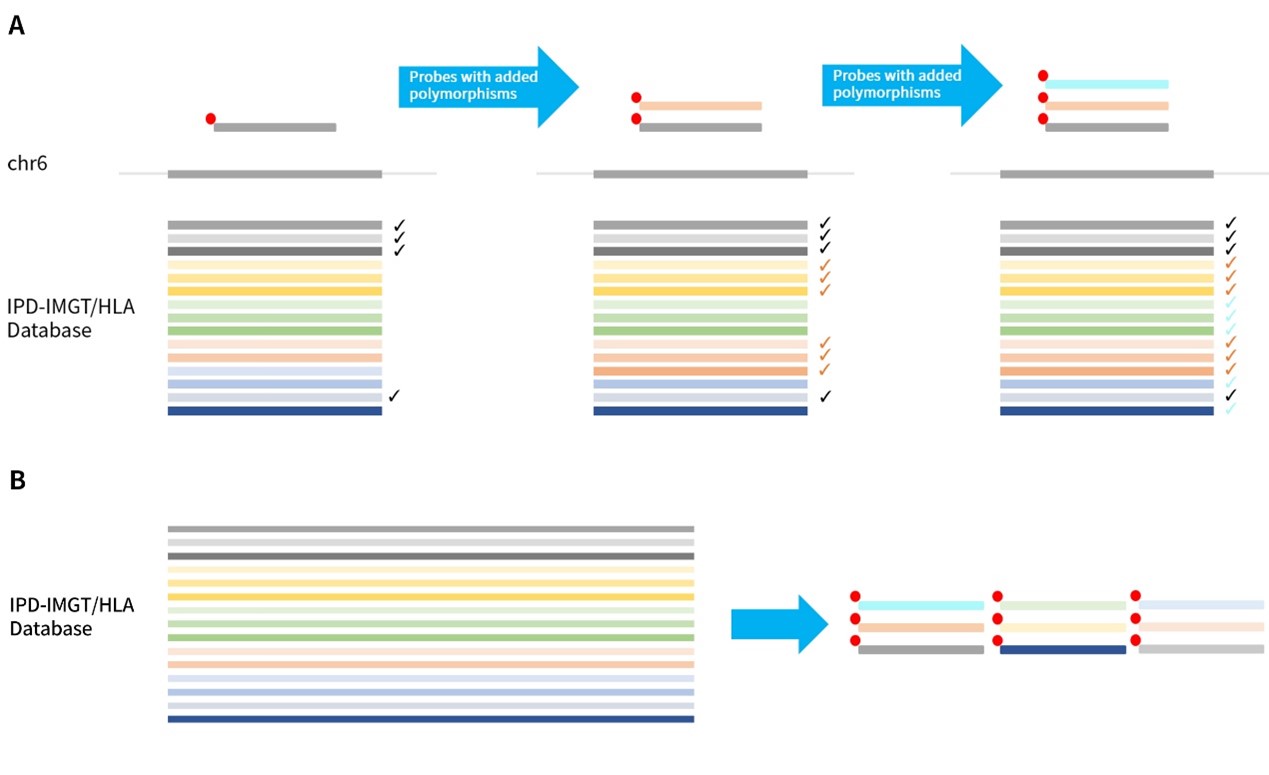
Figure 1. HLA solution from Nanodigmbio. A. HLAtyping Panel v1.0 is based on the reference genome exon probes, supplemented with polymorphic probes according to the HLA database, to achieve effective capture of all types; B. HLAtyping Panel v2.0 can cover all known reference sequences in the latest database (3.50) through polymorphic probe combinations.
The standard for HLA typing is the UCLA DNA Reference Panel (provide the typing reference results to the second or third field), which contains 24 types of Class I and 24 types of Class II.
Libraries were prepared using 50 ng of gDNA and the NadPrep EZ DNA Library Preparation Kit v2, hybrid capture (6 plex) was completed using HLAtyping Panel v2.0, followed by sequencing using NovaSeq 6000 PE150 with the data of approximately 0.5 Gb. Sequencing data were HLA typed using in-house tools.
4.1 Capture performance of HLAtyping Panel v2.0
The target data of the standard is compared to the hg19 reference genome only containing the main chromosome, to evaluate the on-target rate by probe coverage area, and to evaluate the coverage uniformity by the target gene capture area on the main chromosome. The result is shown in Figure 2. (Note: Since the main chromosome genotype of the reference genome is not consistent with each standard, this result only reflects the capture performance of the Panel, and has no direct relationship with the typing performance).
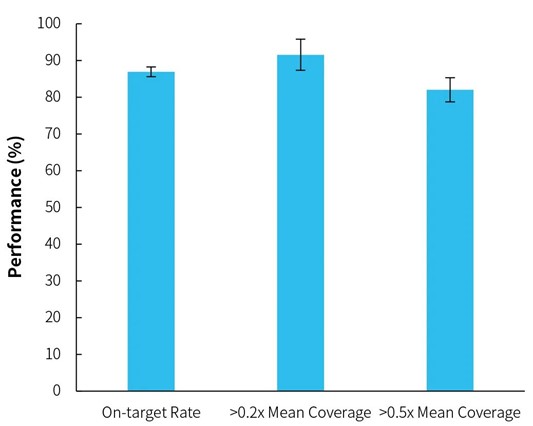
Figure 2. Capture performance of HLAtyping Panel v2.0.
Following targeted capture of standards using HLAtyping Panel v2.0, respectively, sequencing was performed using Illumina® platform.
Note: The standards are Class I and Class II UCLA DNA Reference Panel the capture performance of HLAtyping Panel v2.0 was reflected through calculated mean values and standard deviations of 24 Class I and 24 Class II.
4.2 Genotyping results for Class I by HLAtyping Panel v2.0
Table 1. Genotyping results for Class I by HLAtyping Panel v2.0.
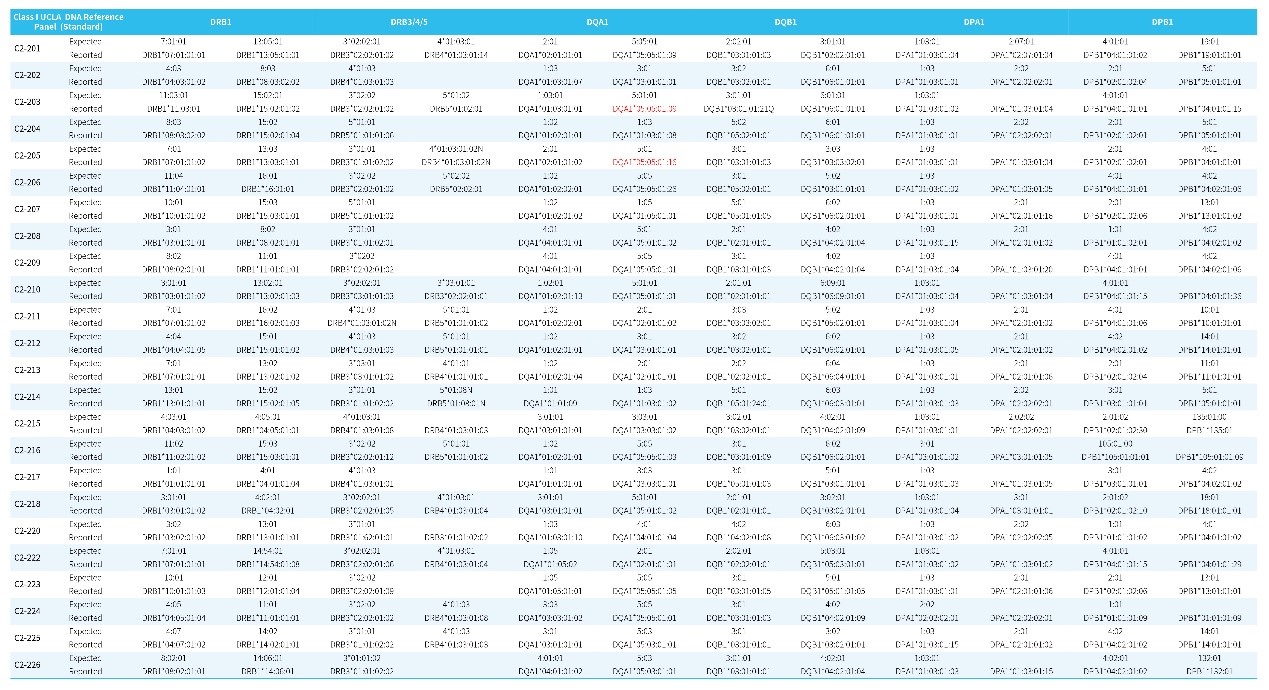
Note: The standard is Class I UCLA DNA Reference Panel. The first row of each standard refers to the typing result for the standard and the second row refers to the data typing result using HLAtyping Panel v2.0.
4.3 Genotyping results for Class II by HLAtyping Panel v2.0
Table 2. Genotyping results for Class II by HLAtyping Panel v2.0.
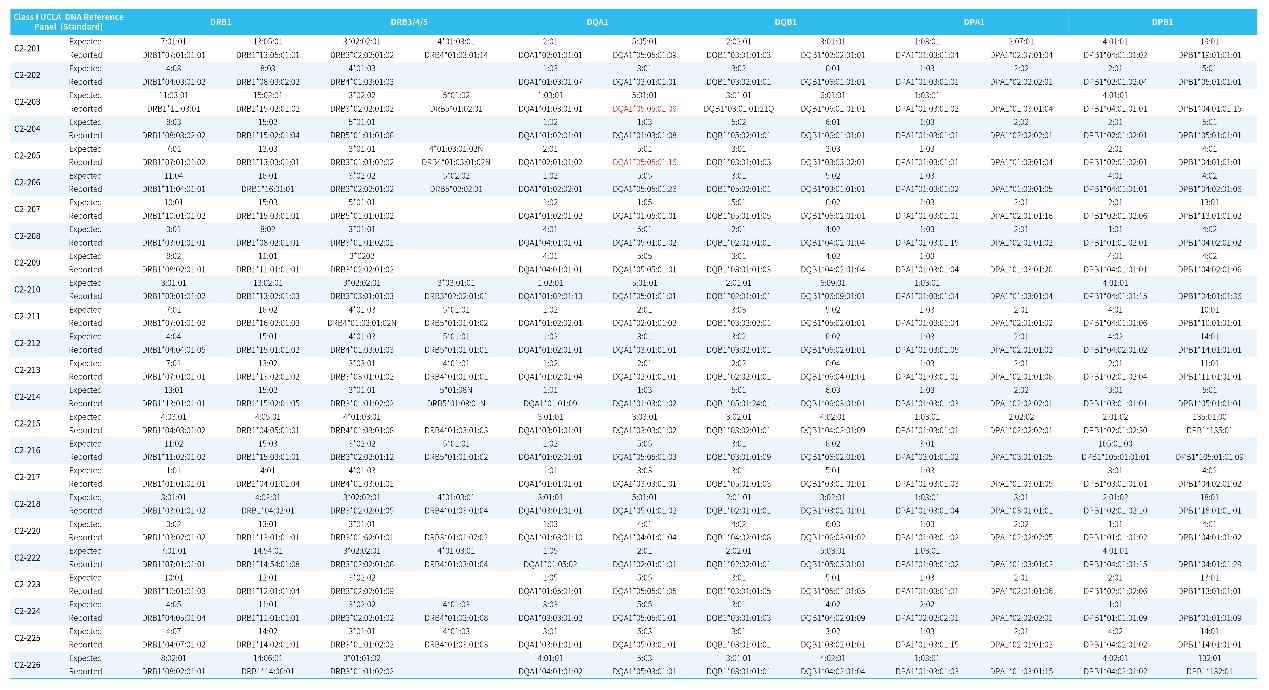
Note: The standard is Class II UCLA DNA Reference Panel. The first row of each standard refers to the typing result for the standard and the second row refers to the data typing result using HLAtyping Panel v2.0.
4.4 Analysis of HLA typing results
The typing results of HLAtyping Panel v2.0 for 24 Class I standard (Table 1.) and 24 Class II standard (Table 2.) are basically consistent with the known results, and the accuracy is higher. For the very few results with differences in the second field, after sequence alignment and inspection, as shown in Figure 3., the new typing results are more consistent: the entire reference sequence showed continuous coverage under the conditions with non-mismatching alignment; while the old results showed interruption of coverage, that is, the sequence composition here lacked of support.
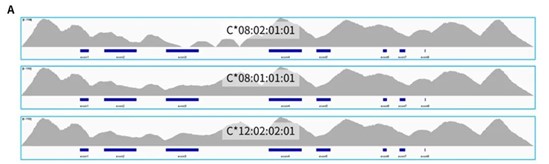
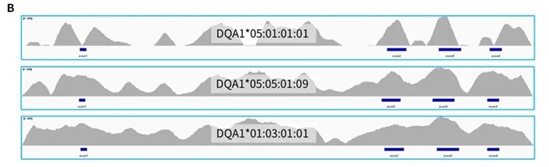
Figure 3. Coverage of reference sequence under non-mismatching conditions with targeted capture using HLAtyping Panel v2.0. A. C*08:02 (top) is the typing result provided by standard C1-213, with discontinuous coverage; C*08:01 (middle) is the new typing result, with continuous coverage; B. DQA1*05:01 top) is the typing result provided by standard C2-203, with discontinuous coverage;DQA1*05:05 (middle) is the new typing result, with continuous coverage; C. DQA1*05:01 ( top) is the typing result provided by standard C2-205, with discontinuous coverage;DQA1*05:05 (middle) is the new typing result, with continuous coverage. All 3 standards are shown the coverage of another allele with consistent results (bottom).
Summary, HLAtyping Panel v2.0 refers to the latest database design, covering the full length of 11 HLA genes, fully utilize the reliability of hybrid capture targeted sequencing technology, and can meet the needs of higher precision HLA typing.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)