How Effective is the Ultra-sensitive Solution for Synchronous Detection of Methylation and Mutation? Let Clinical Samples be the Judge!
View: 2005 / Time: 2024-10-21
Recently, Nanodigmbio launched an ultra-sensitive solution for synchronous detection of methylation and mutation: μCaler™ DNA Full Screen System, and elaborated on the technical principles and unique advantages of this solution through two articles.
Recommended Reading
01 - No Need for Conversion: Ultra-sensitive Solution for Synchronous Detection of Methylation and Mutation in a Single Reaction Coming Soon!
02 - How to Achieve Ultra-sensitive Synchronous Detection of Methylation and Mutation without Sequence Conversion?
In clinical applications, there have always been several challenges in methylation and mutation detection, such as:
• Extremely limited sample amount: Only about 24 ng of cfDNA can be isolated from 10 mL of blood from a healthy individual. How can this precious cfDNA be utilized to detect both markers simultaneously? This presents a major challenge for methodologies and is also the key to improving sensitivity.
• Precarious original information: As the gold standard for DNA methylation detection, the reaction conditions of high temperature and strong alkali in BS conversion often lead to DNA breakage and damage. As a result, the amount of original information that can be preserved post-conversion is extremely limited.
• Time-consuming and labor-intensive operation process: Traditional methods require methylation and mutation detection to be carried out separately, leading to complicated operations that are both time-consuming and labor-intensive.
So, how does μCaler™ DNA Full Screen System perform in clinical samples? Let’s find out!
Application Example
All samples are derived from patients with colorectal cancer. The experimental conditions are as follows:
Traditional methylation detection: Using the NadPrep DNA Methyl Bisulfite Conversion Module to perform BS conversion, followed by pre-library preparation. A Methylation Demo Panel (60 Kb) was then captured using traditional overnight hybridization before sequencing.
Traditional mutation detection: Conventional library were prepared, followed by traditional overnight hybridization capture with NanOnco Panel before sequencing.
μCaler™ DNA Full Screen System for synchronous detection of methylation and mutation: Operate according to the user manual. A custom panel (12 Kb) was designed separately for the synchronous detection of methylation and mutation in colorectal cancer. Methylation sites are selected from those covered by Methylation Demo Panel, while mutation sites were chosen based on high-frequency mutation sites observed in colorectal cancer patients.
Scenario 1: The violent BS conversion reaction often leads to incomplete the information of prepared library and poor stability.
Test results:
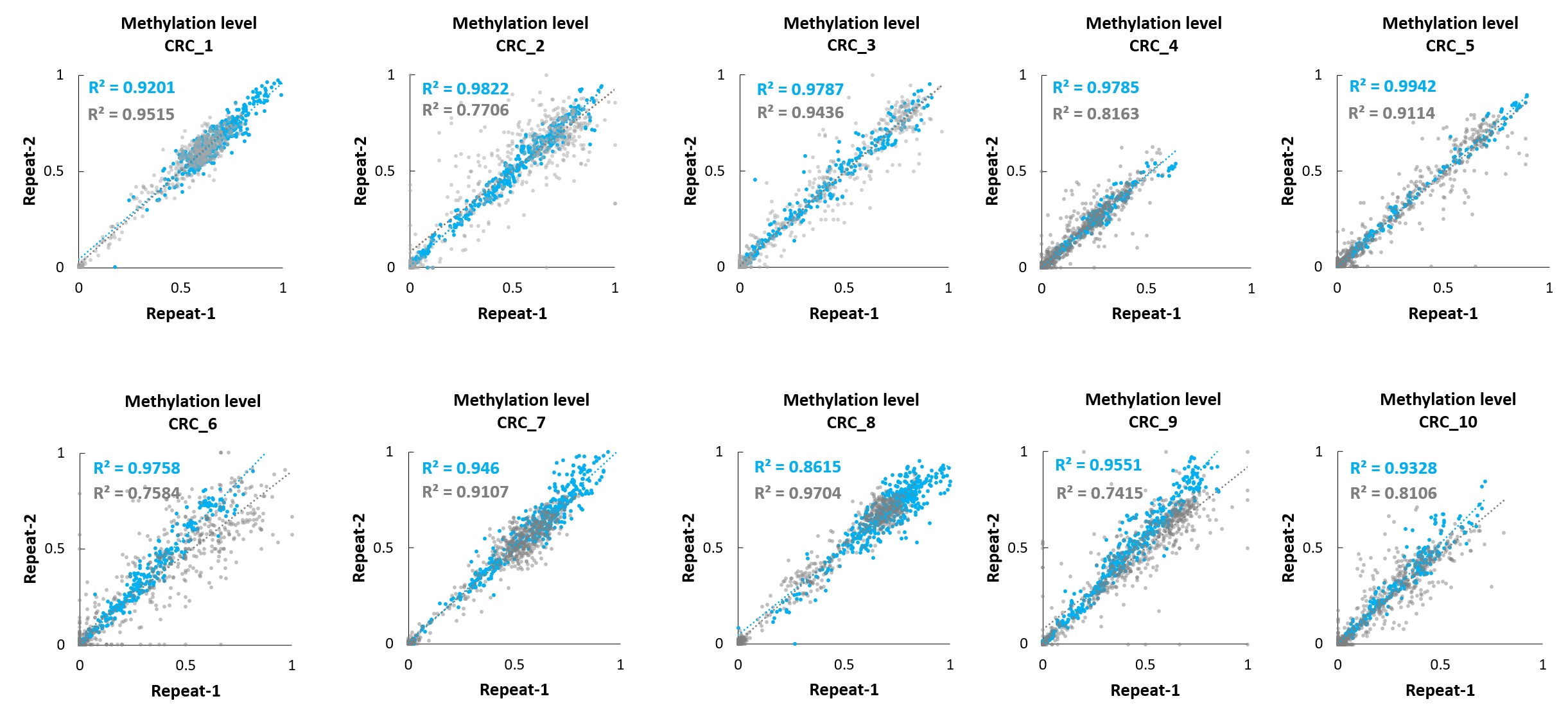
Figure 1. Correlation analysis of methylation levels using different methylation detection routes in tissue samples from 10 colorectal cancer patients.
As shown in Figure 1, the experiment used FFPE tissue samples from 10 colorectal cancer patients. For each sample, 20 ng of DNA was extracted and methylation levels were detected by μCaler™ DNA Full Screen System (blue, abbreviated as Full Screen) and traditional methylation detection (gray, abbreviated as BS), respectively. The results showed that the R² value range of the correlation analysis in the Full Screen group was 0.8615 - 0.9942, while the R² value range of the BS group is 0.7415 - 0.9704. This indicates that in the same sample, the Full Screen group had higher stability in the repeated detection of methylation levels at the same site.
Summary:
Traditional BS treatment is prone to damage and degradation of DNA fragments during the conversion process, which in turn affects the stability of methylation detection results. Therefore, there are certain limitations in applications with low initial amounts of samples. However, Full Screen is based on the MSRE principle and does not require sequence conversion. In theory, it can retain more methylation site information and improve the repeatability and stability of methylation detection results.
Scenario 2: Some clinical samples exhibit both methylation and mutation. Traditional methods require separate detections, resulting in two rounds of pre-library preparation and hybridization, making the process very cumbersome and time-consuming. Can the μCaler™ DNA Full Screen System detect both methylation and mutation in a single round, and how accurate is it?
Test results:
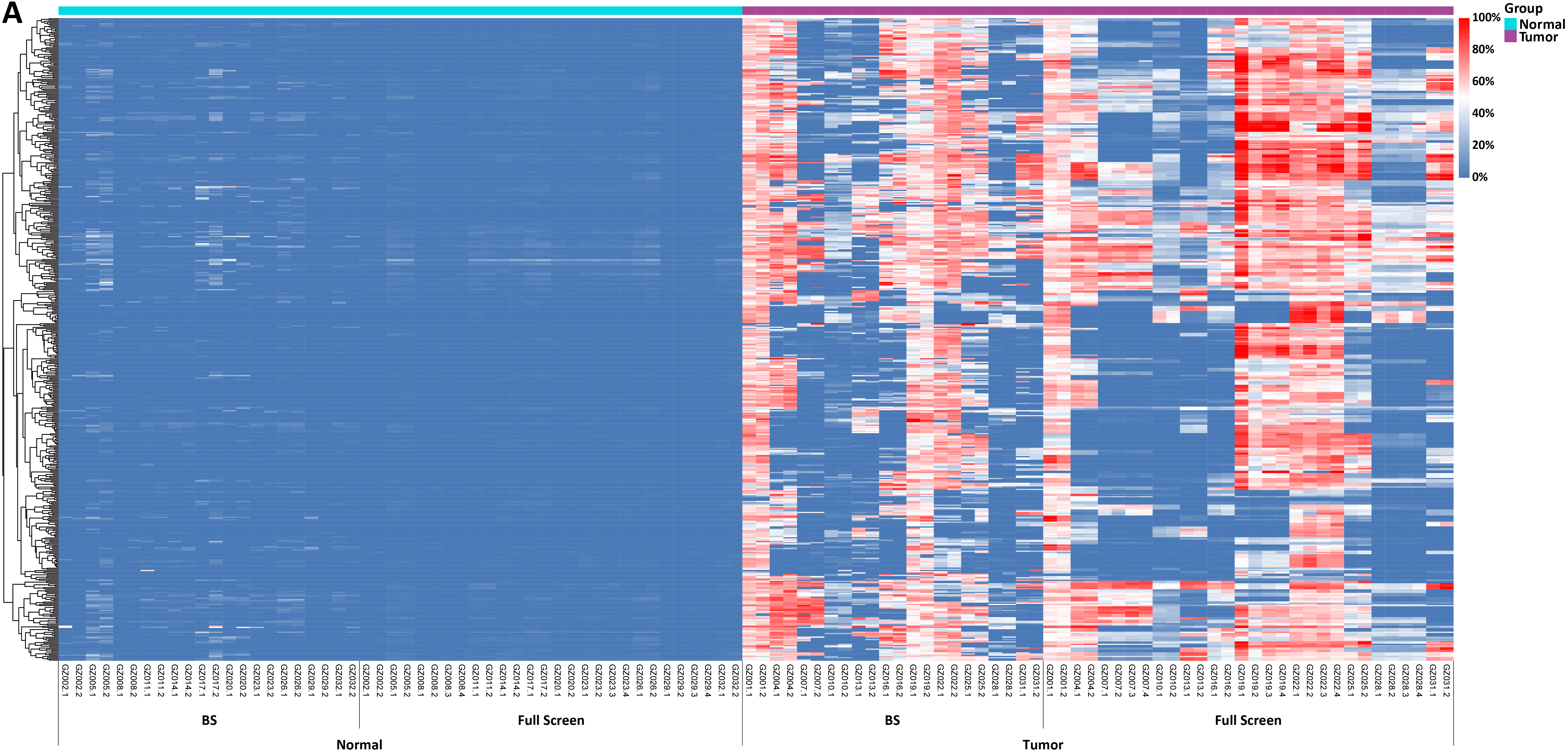
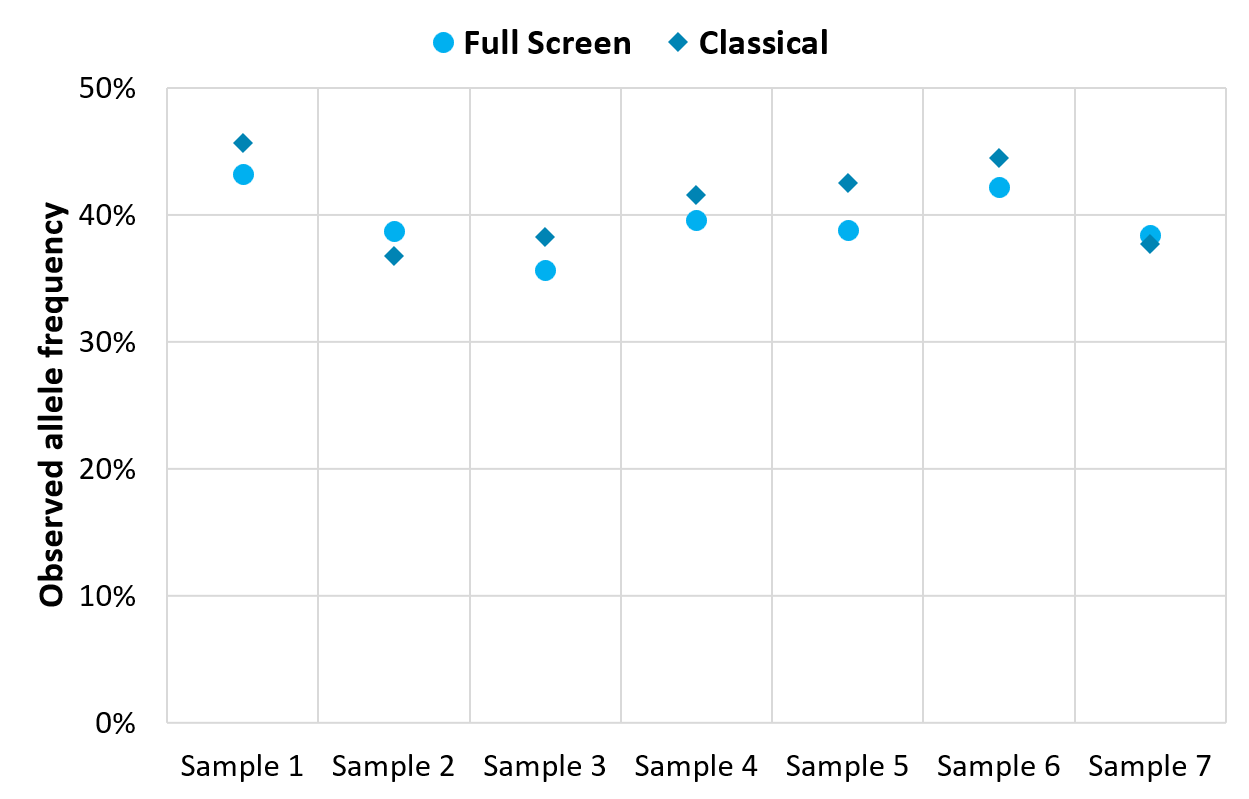
Figure 3. The mutation detection results from Full Screen are consistent with those from traditional mutation detection.
Among these 11 samples of colorectal cancer patients, traditional mutation detection (abbreviated as Classical) identified mutations in 7 samples. For these known mutation sites, the synchronous detection results of Full Screen group were highly consistent with the results of traditional mutation detection (Figure 3.).
Summary:
When dealing with complex tumor samples, Full Screen can not only realize the synchronous detection of methylation and mutation but also significantly increase the coverage depth of CpG sites, and retain more original sequence information, showing its unique advantages in clinical applications.
Scenario 3: I want to explore the potential of this system in early cancer screening. Can it distinguish the methylation levels of cfDNA across different cancer stages?
Test results:
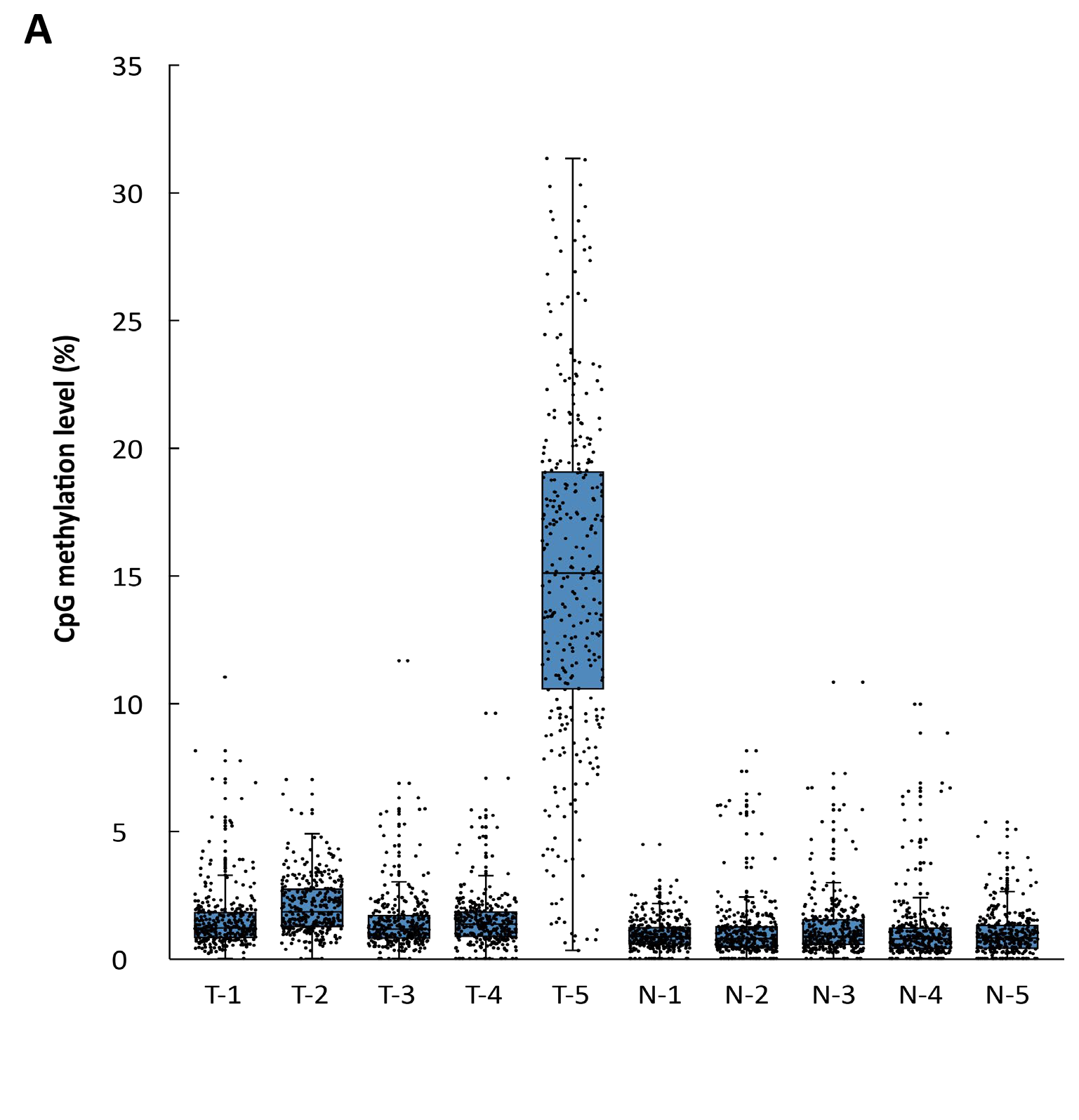
Summary:
Relying on its extremely high sensitivity, μCaler™ DNA Full Screen System can successfully identify methylation signals across different cancer stages, demonstrating its great potential as a tool for cancer screening and early diagnosis. By detecting changes in methylation and mutation signals earlier and implementing timely interventions, this technology is expected to help improve the success rate of cancer treatment and the prognosis of patients.
Scenario 4: The operation time of the traditional hybridization capture method exceeds 40 hours and is expensive. How to reduce costs?
Traditional hybridization capture method involve separate detection of methylation and mutation biomarkers, requiring two separate input samples and two sets of capture probes for two hybridization reactions. The total time-consuming process before sequencing is as long as more than 46 hours, with extremely high time costs. However, Full Screen greatly simplifies the operation process by combining the dual advantages of MSRE and the ultra-sensitive probe capture technology μCaler™ based on Nanodigmbios' exclusive patent. Only one initial input sample and one set of capture probe for one hybridization reaction, are needed to achieve ultra-high sensitivity synchronous detection of methylation and panoramic mutation. This significantly reduces the testing time, with the entire process completed within 8 hours. At the same time, Full Screen offers powerful technical support for early screening, early diagnosis, precise treatment and prognosis evaluation of tumors.
Recommended Reading
01 - No Need for Conversion: Ultra-sensitive Solution for Synchronous Detection of Methylation and Mutation in a Single Reaction Coming Soon!
02 - How to Achieve Ultra-sensitive Synchronous Detection of Methylation and Mutation without Sequence Conversion?
In clinical applications, there have always been several challenges in methylation and mutation detection, such as:
• Extremely limited sample amount: Only about 24 ng of cfDNA can be isolated from 10 mL of blood from a healthy individual. How can this precious cfDNA be utilized to detect both markers simultaneously? This presents a major challenge for methodologies and is also the key to improving sensitivity.
• Precarious original information: As the gold standard for DNA methylation detection, the reaction conditions of high temperature and strong alkali in BS conversion often lead to DNA breakage and damage. As a result, the amount of original information that can be preserved post-conversion is extremely limited.
• Time-consuming and labor-intensive operation process: Traditional methods require methylation and mutation detection to be carried out separately, leading to complicated operations that are both time-consuming and labor-intensive.
So, how does μCaler™ DNA Full Screen System perform in clinical samples? Let’s find out!
Application Example
All samples are derived from patients with colorectal cancer. The experimental conditions are as follows:
Traditional methylation detection: Using the NadPrep DNA Methyl Bisulfite Conversion Module to perform BS conversion, followed by pre-library preparation. A Methylation Demo Panel (60 Kb) was then captured using traditional overnight hybridization before sequencing.
Traditional mutation detection: Conventional library were prepared, followed by traditional overnight hybridization capture with NanOnco Panel before sequencing.
μCaler™ DNA Full Screen System for synchronous detection of methylation and mutation: Operate according to the user manual. A custom panel (12 Kb) was designed separately for the synchronous detection of methylation and mutation in colorectal cancer. Methylation sites are selected from those covered by Methylation Demo Panel, while mutation sites were chosen based on high-frequency mutation sites observed in colorectal cancer patients.
Scenario 1: The violent BS conversion reaction often leads to incomplete the information of prepared library and poor stability.
Test results:

Figure 1. Correlation analysis of methylation levels using different methylation detection routes in tissue samples from 10 colorectal cancer patients.
As shown in Figure 1, the experiment used FFPE tissue samples from 10 colorectal cancer patients. For each sample, 20 ng of DNA was extracted and methylation levels were detected by μCaler™ DNA Full Screen System (blue, abbreviated as Full Screen) and traditional methylation detection (gray, abbreviated as BS), respectively. The results showed that the R² value range of the correlation analysis in the Full Screen group was 0.8615 - 0.9942, while the R² value range of the BS group is 0.7415 - 0.9704. This indicates that in the same sample, the Full Screen group had higher stability in the repeated detection of methylation levels at the same site.
Summary:
Traditional BS treatment is prone to damage and degradation of DNA fragments during the conversion process, which in turn affects the stability of methylation detection results. Therefore, there are certain limitations in applications with low initial amounts of samples. However, Full Screen is based on the MSRE principle and does not require sequence conversion. In theory, it can retain more methylation site information and improve the repeatability and stability of methylation detection results.
Scenario 2: Some clinical samples exhibit both methylation and mutation. Traditional methods require separate detections, resulting in two rounds of pre-library preparation and hybridization, making the process very cumbersome and time-consuming. Can the μCaler™ DNA Full Screen System detect both methylation and mutation in a single round, and how accurate is it?
Test results:

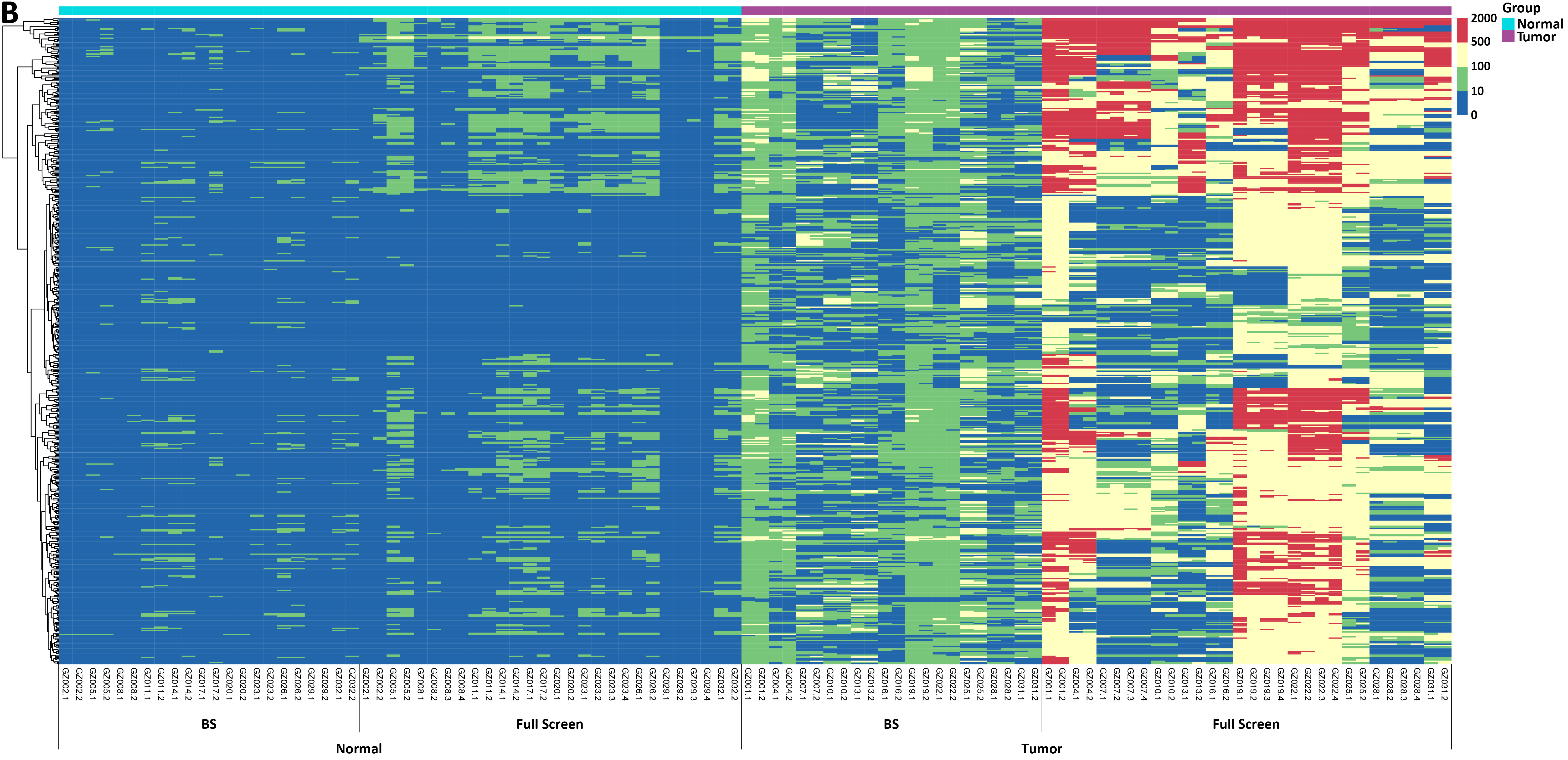
Figure 2. Methylation levels and depth of each CpG site in tissue samples of 11 colorectal cancer patients. A. Methylation levels of CpG sites; B. Coverage depth of methylated CpG sites.
In another test, tissue samples from 11 colorectal cancer patients were used to detect the methylation levels of each CpG site by Full Screen and BS methods respectively. The results showed significant differences between tumor samples and normal samples, with both methods effectively distinguishing between tumor and normal samples (Figure 2. A). In addition, when analyzing the coverage depth of methylated CpG sites, it was found that the average depth of methylated CpG sites in the tumor sample reached 252.7x in Full Screen group, while the average depth of the BS group could only reach 10.8x (Figure 2. B).
Figure 3. The mutation detection results from Full Screen are consistent with those from traditional mutation detection.
Among these 11 samples of colorectal cancer patients, traditional mutation detection (abbreviated as Classical) identified mutations in 7 samples. For these known mutation sites, the synchronous detection results of Full Screen group were highly consistent with the results of traditional mutation detection (Figure 3.).
Summary:
When dealing with complex tumor samples, Full Screen can not only realize the synchronous detection of methylation and mutation but also significantly increase the coverage depth of CpG sites, and retain more original sequence information, showing its unique advantages in clinical applications.
Scenario 3: I want to explore the potential of this system in early cancer screening. Can it distinguish the methylation levels of cfDNA across different cancer stages?
Test results:

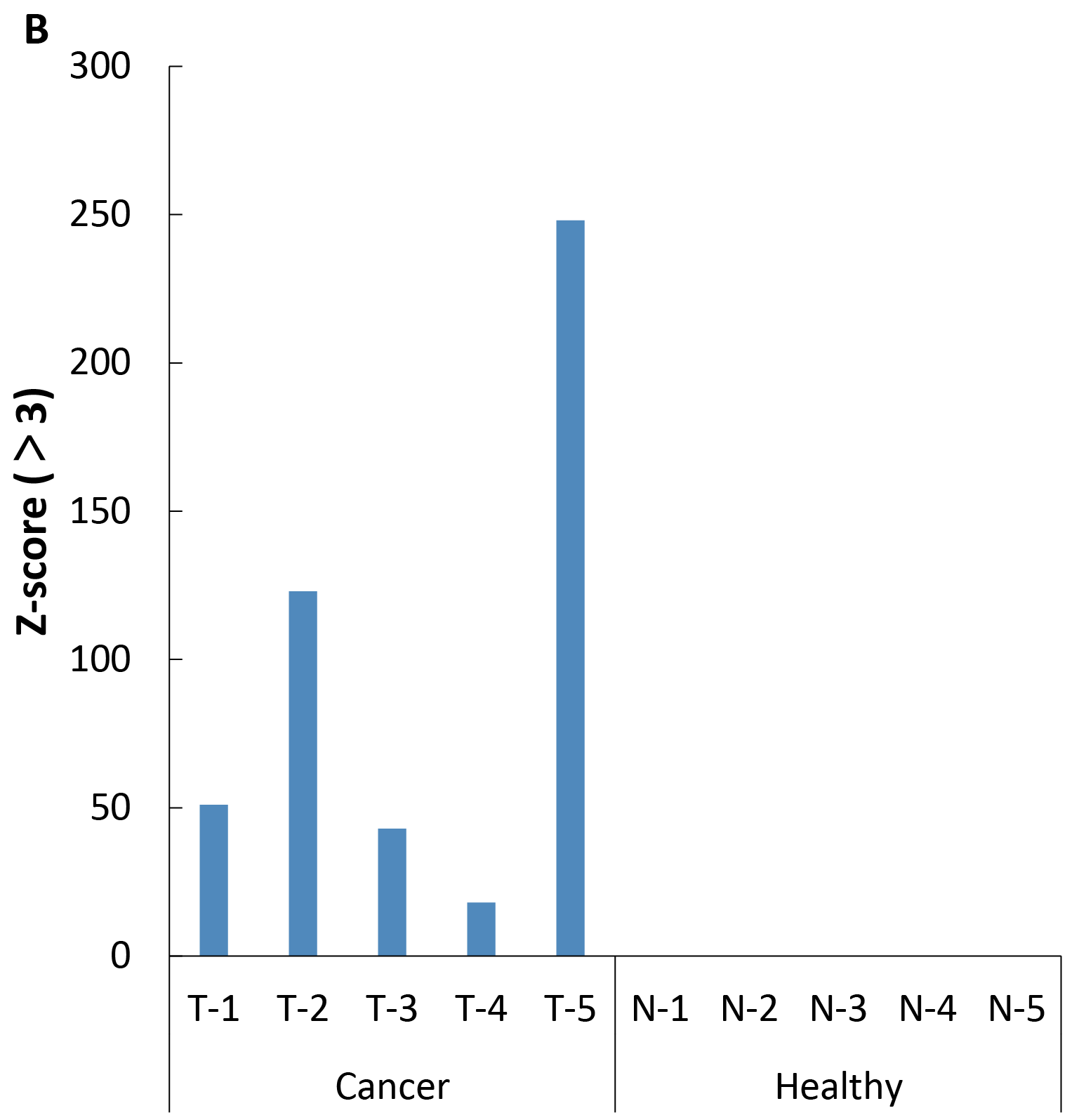
Figure 4. A. Methylation levels of CpG sites; B. Statistical results with Z - score > 3.
After extracting cfDNA from the plasma of 5 colorectal cancer patients at different stages (T1 - T5) and 5 healthy individuals (N1 - N5), with initial input amounts ranging from 2 to 10 ng, the methylation levels were detected by Full Screen. As shown in Figure 4, the results indicated that Full Screen can clearly distinguish early stage cancer patients (T1 - T4), late stage cancer patients (T5) and healthy individuals.Summary:
Relying on its extremely high sensitivity, μCaler™ DNA Full Screen System can successfully identify methylation signals across different cancer stages, demonstrating its great potential as a tool for cancer screening and early diagnosis. By detecting changes in methylation and mutation signals earlier and implementing timely interventions, this technology is expected to help improve the success rate of cancer treatment and the prognosis of patients.
Scenario 4: The operation time of the traditional hybridization capture method exceeds 40 hours and is expensive. How to reduce costs?
Traditional hybridization capture method involve separate detection of methylation and mutation biomarkers, requiring two separate input samples and two sets of capture probes for two hybridization reactions. The total time-consuming process before sequencing is as long as more than 46 hours, with extremely high time costs. However, Full Screen greatly simplifies the operation process by combining the dual advantages of MSRE and the ultra-sensitive probe capture technology μCaler™ based on Nanodigmbios' exclusive patent. Only one initial input sample and one set of capture probe for one hybridization reaction, are needed to achieve ultra-high sensitivity synchronous detection of methylation and panoramic mutation. This significantly reduces the testing time, with the entire process completed within 8 hours. At the same time, Full Screen offers powerful technical support for early screening, early diagnosis, precise treatment and prognosis evaluation of tumors.
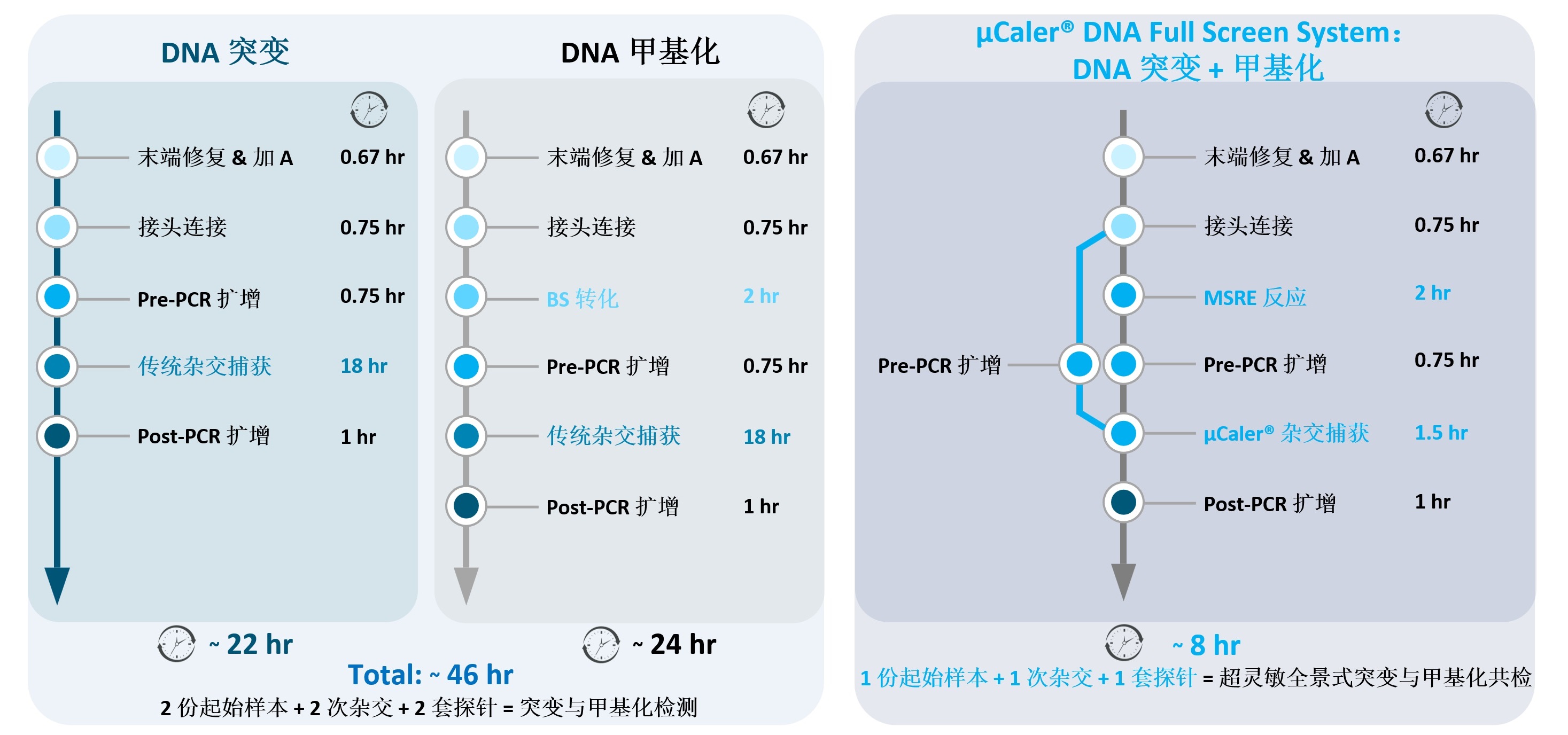
Figure 5. Comparison of workflows and time consumption for different dual-biomarkers detection methods for methylation and mutation.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)