Can 768 Index Combinations Meet the Growing High-throughput Demands of Various Sequencers?
With the rapid advancements in genomics, transcriptomics, and single-cell sequencing technologies, the demand for high-throughput sequencing in biological-related field has surged dramatically. This is particularly evident in cutting-edge areas such as precision medicine, infectious disease research, and environmental ecology. This growing demand has made sequencing throughput a core focus in the development of sequencing technologies, prompting major sequencer manufacturers to compete in delivering higher data output at lower costs.
For example, Illumina’s NovaSeq X/X Plus sequencing systems launched in 2022 offers an impressive sequencing throughput of 16 Tb. Meanwhile, MGI’s newly released DNBSEQ-T20×2 sequencing system achieves an even more astonishing throughput of 72 Tb. To put this into perspective, the NovaSeq X Plus, with its 16 Tb throughput, can process 178 samples at 30x with whole genome sequencing (WGS) or 800 samples at 250x with whole exome sequencing (WES), significantly enhancing sequencing efficiency.
Index sequences, as unique markers, are essential for demultiplexing data from different samples after sequencing. They prevent sample misassignment and cross-contamination, ensuring the accuracy and reliability of sequencing results. Nanodigmbio previously launched the NadPrep Universal Stubby Adapter (UDI) Module, a patented dual-platform (Illumina & MGI) adapter module featuring 384 Index combinations. Libraries prepared with this module not only support the Illumina sequencing platforms but can also be directly circularized for use on the MGI sequencing platforms without App-A conversion, saving valuable time and earning widespread praise from users. Now, Nanodigmbio has expanded and upgraded the combinations of indexes in this module to 768, following strict quality control and meticulous optimization. This enhancement aims to better meeting the growing demand for higher throughput in the market.
02 Nanodigmbio’s Exclusive Adapter Design
NadPrep Universal Stubby Adapter (UDI) Module contains universal stubby adapters and 10 nt Unique Dual Index amplification primers. It can be used for preparing libraries with Illumina structures and 5' phosphorylation modification; the libraries can be sequenced on Illumina platform directly and on MGI platform after circularization without App-A conversion.
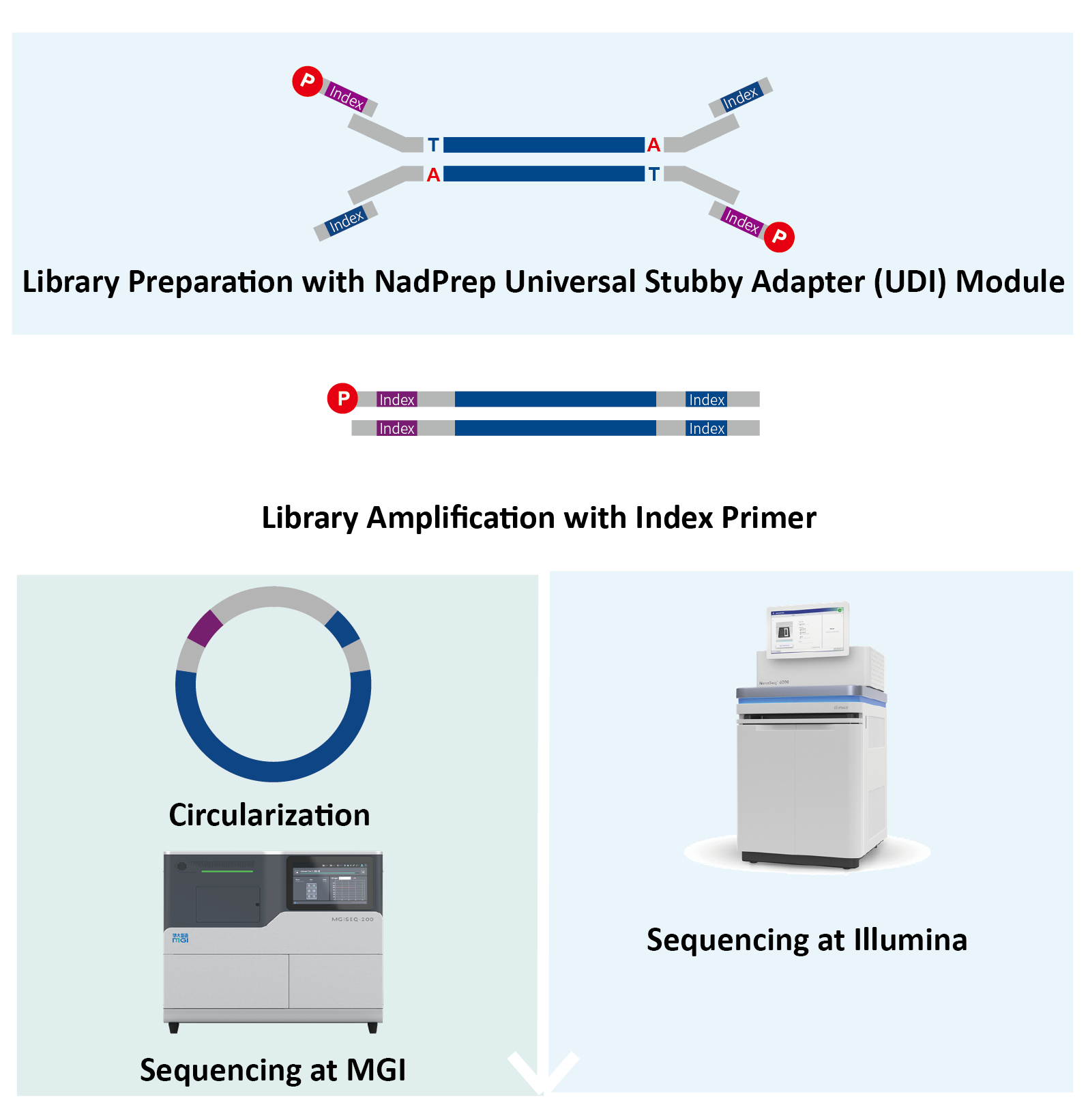
Nanodigmbio has expanded the Index combinations in the NadPrep Universal Stubby Adapter (UDI) Module from 384 to 768, with Indexes 385 - 768 designed to meet the 8 Index balance principle. This ensures compliance with both the 8-base, 8-Index minimum color balance required by Illumina platforms and the 10-base, 8-Index minimum color balance required by MGI platforms. Additionally, the module’s packaging design is also user-friendly, employing a 96-well plate format with single-reaction quantities per well. This reduces contamination risks and enhances the experimental convenience. Furthermore, this design is compatible for NGS automation workstations, enabling efficient laboratory operations.
03 Performance
3.1 Advantages of 8-Index balanced design
The newly expanded Indexes 385 - 768 in the NadPrep Universal Stubby Adapter (UDI) Module feature an 8-Index balanced design, enabling compliance with the base balance rules for both 8 nt and 10 nt sequencing modes. This supports diverse multiplexed sequencing scenarios. When using ≥ 8 consecutive Index libraries with proportional mixing, each base in the Index satisfies the ≥ 12.5% balance ratio recommended for MGI sequencing platforms. In contrast, some adapter designs targeting Illumina platforms fail to fully consider base balance principles, making them unable to meet sequencing requirements, even with up to 14 consecutive Indexes (Figure 2.).
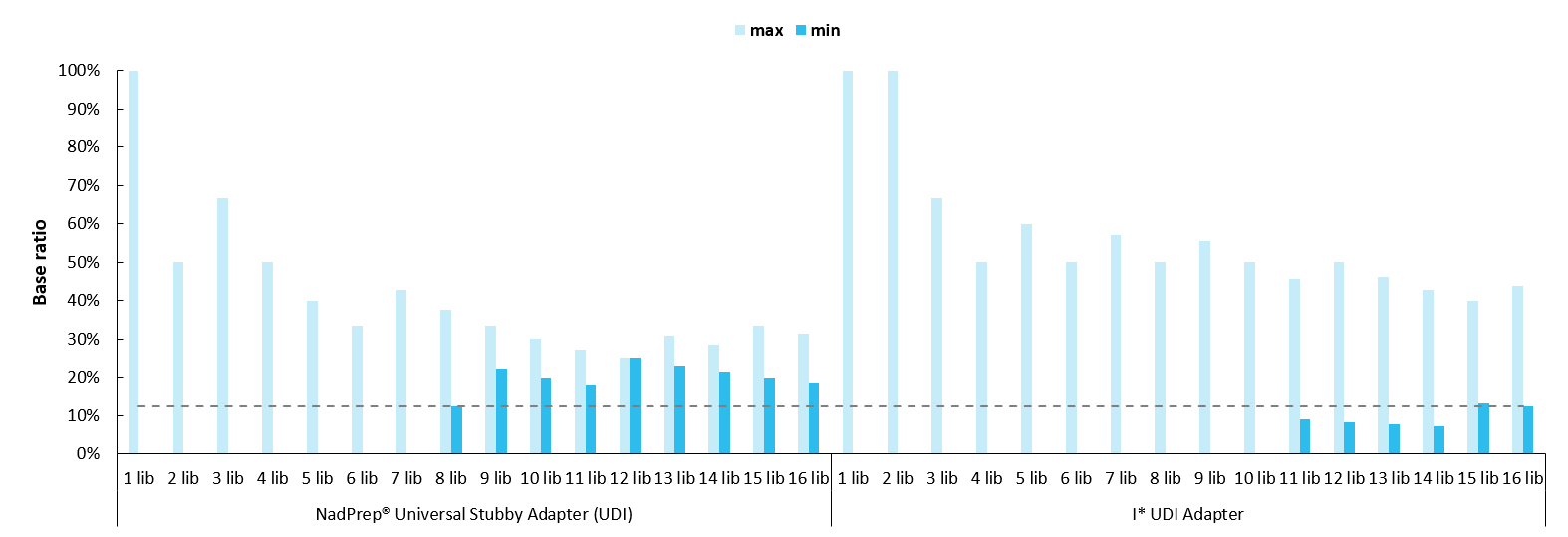
Figure 2. Advantages of 8-Index balanced design in NadPrep Universal Stubby Adapter (UDI) Module (Index 385 - 768).
3.2 Stable and Efficient Library Yield
Using 10 ng of Human Genomic DNA (Promega, G1471), WGS libraries were prepared with NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep Universal Stubby Adapter (UDI) Module (Index 385-768), amplified for 9 cycles. The results showed that the average library yield of 2173.4 ng (1865 ng - 2465 ng) across 384 WGS libraries (Figure 3.).
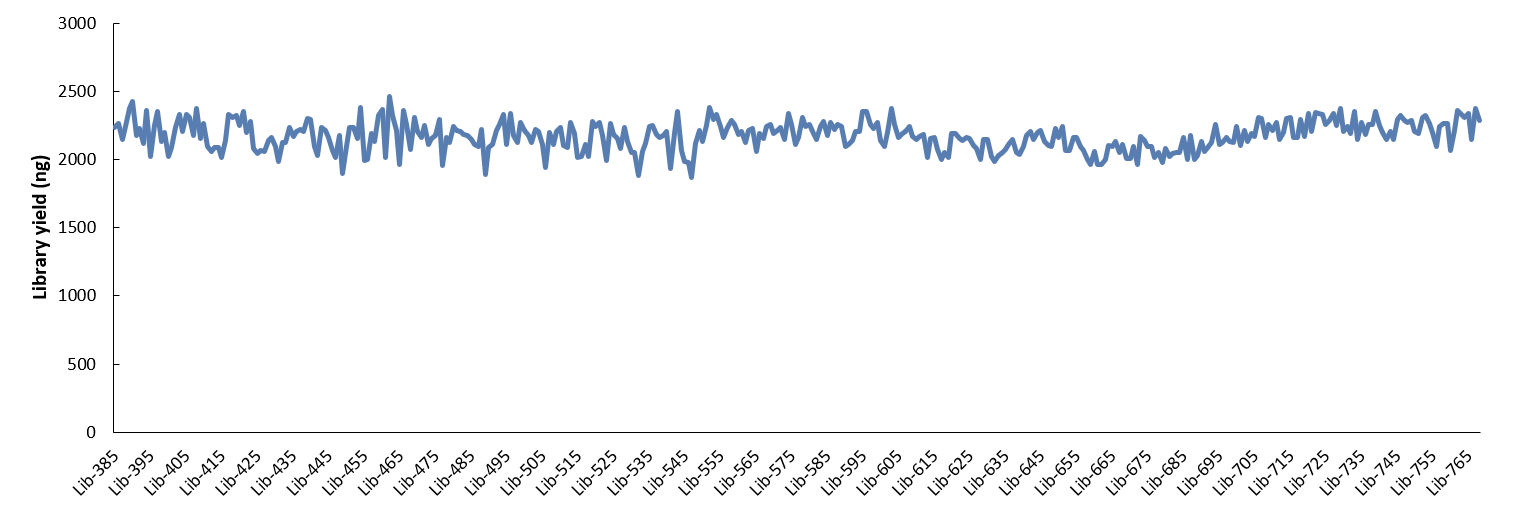
Figure 3. Efficient and stable library yield with NadPrep Universal Stubby Adapter (UDI) Module (Index 385 - 768).
3.3 Uniform Effective Data Splitting Rate
The pre-libraries prepared in step 3.2 were mixed in equal proportions, and portions of the mixed libraries were used for data splitting tests in three dimensions: WGS_MGI, WGS_Illumina, and Hyb_Illumina.
• WGS_MGI: The mixed pre-libraries were circularized using NadPrep Universal Circularization Kit v2, and then sequenced on the MGI platform (DNBSEQ-T7, PE150; 0.1 Gb/Lib).
• WGS_Illumina: The mixed pre-libraries were directly sequenced on the Illumina platform (Novaseq 6000, PE150; 0.1 Gb/Lib).
• Hyb_Illumina: The mixed pre-libraries were performed for hybridization capture using NadPrep Hybrid Capture Reagents and LungCancer Panel v1.0, followed by sequencing on the Illumina platform (Novaseq 6000, PE150; 0.1 Gb/Lib).
Data analysis revealed highly uniform effective splitting rates across all experimental and sequencing setups using the newly expanded Indexes 385 - 768 (Figure 4.).
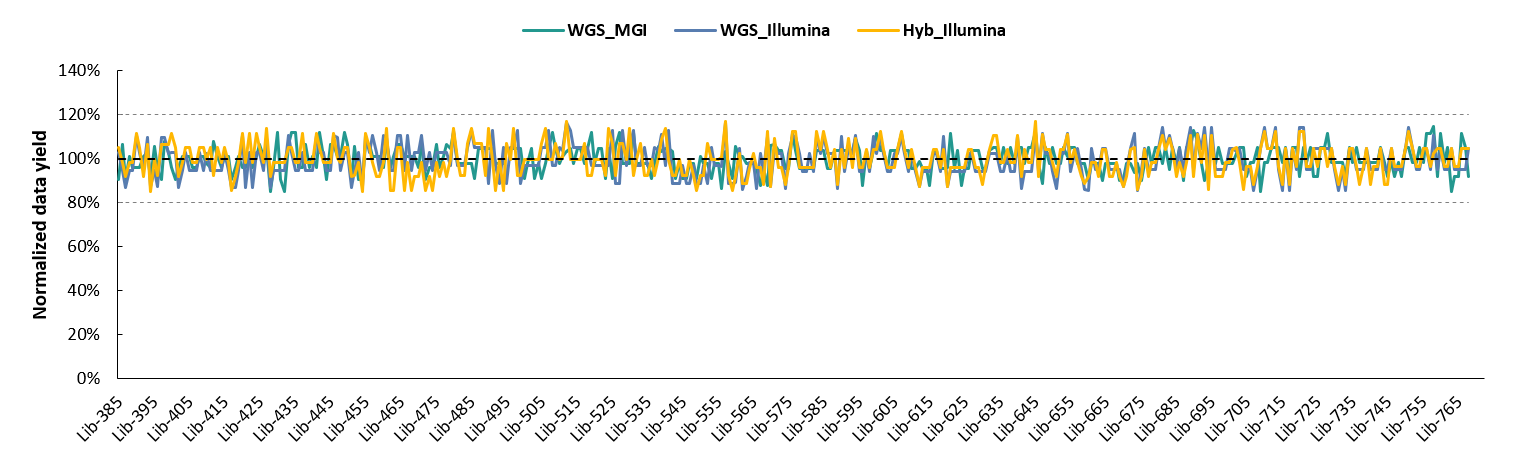
Figure 4. Highly uniform effective splitting rates across different library types with NadPrep Universal Stubby Adapter (UDI) Module (Index 385 - 768).
3.4 Significant Improvement in Data Accuracy
A mixed cross-contamination test was performed on libraries prepared with NadPrep Universal Stubby Adapter (UDI) Module (Index 385 - 768). The results showed that all Index sequences were correctly split, with the perfect match paired barcode percentage exceeding 98.5%, significantly improving sequencing accuracy and effectively reducing data errors caused by Index cross-contamination during sequencing.
Figure 5. Cross-contamination analysis of libraries prepared with NadPrep Universal Stubby Adapter (UDI) Module (Index 385 - 768). WGS libraries with 384 Indexes were mixed in a single lane for sequencing.
Note: Green color represents perfect match paired barcode percentage > 98.5%.Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)